
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,678,383 – 18,678,517 |
| Length | 134 |
| Max. P | 0.898443 |

| Location | 18,678,383 – 18,678,477 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.56 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18678383 94 + 22224390 ------AGCUGCUCU--CGUUAACUU------------------AUUCAAAACUGAAAUUUCUUUAUCAUUUCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGU ------....(((((--((((.....------------------.((((....))))..........(((((....))))))))))...(((((((.((((....))))))))))))))) ( -27.70) >DroSim_CAF1 3925 94 + 1 ------AGGUGCUCU--CGUCAACUU------------------AUCUAAAUCUGGAAUUCCUUUGUCUUUCCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGU ------....(((((--((((.....------------------........((((((...........))))))(....))))))...(((((((.((((....))))))))))))))) ( -31.60) >DroYak_CAF1 3748 117 + 1 AGCCCAAGCUGGUCCGCCUCU-GUUUGCCAAAGUGAAUAUGCUCAUCCAAAUCUGGAACU-CUUUGUCU-UUCAGCAAAUGGACGAUCCGCCAUCGCUGCCCGUGGGCACGGAGGUAAGU (((....)))((..((..(((-((((((((((((((......)))((((....))))...-)))))...-....)))))))))))..))(((..((.((((....))))))..))).... ( -32.31) >consensus ______AGCUGCUCU__CGUUAACUU__________________AUCCAAAUCUGGAAUU_CUUUGUCUUUUCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGU .............................................((((...((((((...........))))))....)))).....((((((((.((((....))))))))))))... (-23.62 = -23.73 + 0.12)


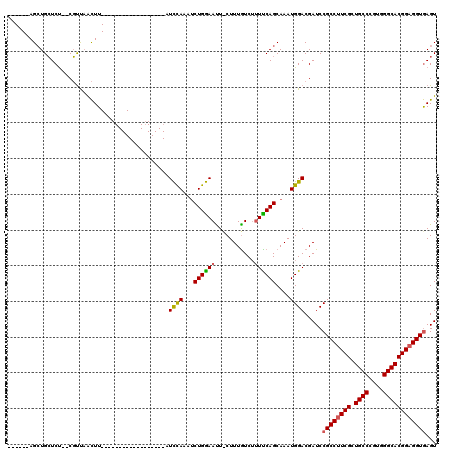
| Location | 18,678,401 – 18,678,517 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -39.12 |
| Energy contribution | -38.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
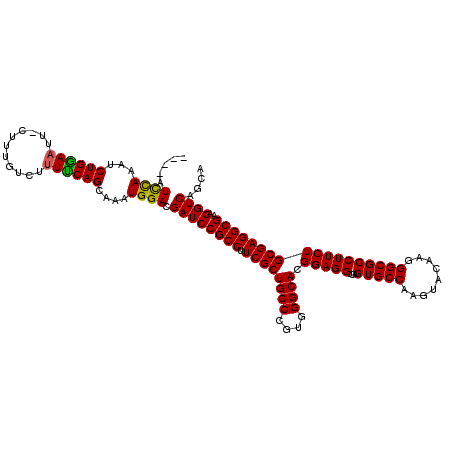
>X_DroMel_CAF1 18678401 116 + 22224390 ----AUUCAAAACUGAAAUUUCUUUAUCAUUUCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGUGCCAAGUACAAGGGCGCCUUCUGCGAGGCGAAGGUCAGCA ----.((((...(((((((.........)))))))....)))).((((((((((((.((((....))))))))))))..((((..(((.((((...)))).)))..))))..)))).... ( -39.80) >DroSim_CAF1 3943 116 + 1 ----AUCUAAAUCUGGAAUUCCUUUGUCUUUCCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGUGCCAAGUACAAGGGCGCCUUCUGCGAGGCGAAGGUCAGCA ----........((((....((((((((((....(((...((.((.((((((((((.((((....)))))))))))..((((...))))..)))))))...))))))))))))))))).. ( -44.00) >DroYak_CAF1 3787 118 + 1 GCUCAUCCAAAUCUGGAACU-CUUUGUCU-UUCAGCAAAUGGACGAUCCGCCAUCGCUGCCCGUGGGCACGGAGGUAAGUGCCAAGUACAAGGGCGCCUUCUGCGAGGCGAAGGUCAGCA (((..((((....))))..(-((((((((-(.(((.....((.((.((((((..((.((((....))))))..)))..((((...))))..)))))))..))).))))))))))..))). ( -42.40) >consensus ____AUCCAAAUCUGGAAUU_CUUUGUCUUUUCAGCAAAUGGACGAUCCGCCUUCGCUGCCCGUGGGCACGGAGGUGAGUGCCAAGUACAAGGGCGCCUUCUGCGAGGCGAAGGUCAGCA .....((((...((((((...........))))))....)))).((((((((.((((((((....)))).(((((...(((((.........))))))))))))))))))..)))).... (-39.12 = -38.57 + -0.55)

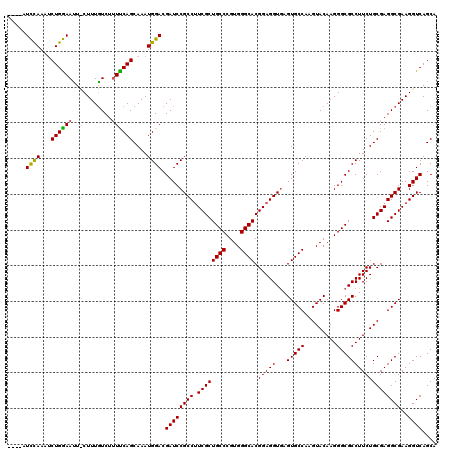
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:12 2006