
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,677,486 – 18,677,579 |
| Length | 93 |
| Max. P | 0.991523 |

| Location | 18,677,486 – 18,677,579 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
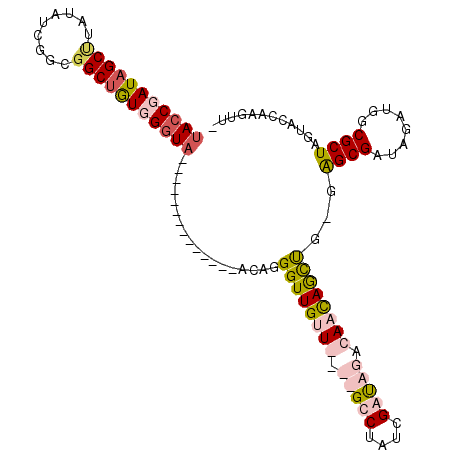
>X_DroMel_CAF1 18677486 93 + 22224390 -AACUUGGUAUUAGCGCCAUCUGGCGCUCUCAGCUGUUGCCUAUCGAUAGGC----AACAACCCUAU--------------UACCAACAGCCGCCGAUAUAGGCUAUCGGUA -...((((((..((((((....))))))......(((((((((....)))))----)))).......--------------)))))).....(((((((.....))))))). ( -36.90) >DroSim_CAF1 3005 93 + 1 -AACUUGCUACUAGCGCCAUCUACCGCUCCCAGCUGUUGUCUAUCGAUAGGC----AACAACCUUAU--------------UAUCAAUAGCCGCCGAUAUACGCUAUCGGUA -.....(((...((((........))))...)))(((((((((....)))))----)))).......--------------...........(((((((.....))))))). ( -24.30) >DroEre_CAF1 4707 111 + 1 CAACUUCGUACUAGCGCCAUCUAUCGCCC-CAGUUGGUGUAUGUCGAUAGACAUCCAACAACCCUGUUUCACACACAUUCUUACCCACUGCCGCCGAUAGGAGCUAUCGGUA .....(((...((((....((((((((..-((((.((((.(((((....)))))..((((....)))).............)))).))))..).))))))).)))).))).. ( -26.20) >DroYak_CAF1 2713 112 + 1 CUACUUGCCACUAGCGCCAUCUAUCGCCCGCGACUAGUGUAUAUCGAUAGACAUCCAACAACCCUGUUUCCUCAACAUUCUGACCCACAGCCGCCGAUAAAAGCUAUCGGUA ........(((((((((............))).))))))..(((((((((.............((((....(((......)))...)))).............))))))))) ( -23.07) >consensus _AACUUGCUACUAGCGCCAUCUAUCGCCC_CAGCUGGUGUAUAUCGAUAGAC____AACAACCCUAU______________UACCAACAGCCGCCGAUAUAAGCUAUCGGUA .............(((........)))......((((((.....))))))..........................................(((((((.....))))))). (-13.25 = -13.38 + 0.12)


| Location | 18,677,486 – 18,677,579 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.43 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -18.35 |
| Energy contribution | -20.85 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18677486 93 - 22224390 UACCGAUAGCCUAUAUCGGCGGCUGUUGGUA--------------AUAGGGUUGUU----GCCUAUCGAUAGGCAACAGCUGAGAGCGCCAGAUGGCGCUAAUACCAAGUU- (((((((((((.........)))))))))))--------------...((((((((----(((((....)))))))))))....((((((....))))))....)).....- ( -46.20) >DroSim_CAF1 3005 93 - 1 UACCGAUAGCGUAUAUCGGCGGCUAUUGAUA--------------AUAAGGUUGUU----GCCUAUCGAUAGACAACAGCUGGGAGCGGUAGAUGGCGCUAGUAGCAAGUU- ..((((((.....)))))).(.((((((((.--------------...((((....----)))))))))))).)....((((..((((.(....).))))..)))).....- ( -28.90) >DroEre_CAF1 4707 111 - 1 UACCGAUAGCUCCUAUCGGCGGCAGUGGGUAAGAAUGUGUGUGAAACAGGGUUGUUGGAUGUCUAUCGACAUACACCAACUG-GGGCGAUAGAUGGCGCUAGUACGAAGUUG ..((((((((((.((((.((....)).))))......(((.....)))))))))))))(((((....)))))......((((-(.((........)).)))))......... ( -31.60) >DroYak_CAF1 2713 112 - 1 UACCGAUAGCUUUUAUCGGCGGCUGUGGGUCAGAAUGUUGAGGAAACAGGGUUGUUGGAUGUCUAUCGAUAUACACUAGUCGCGGGCGAUAGAUGGCGCUAGUGGCAAGUAG ..(((((((((((..((((((.(((.....)))..))))))(....))))))))))))(((((....)))))..(((.(((((.((((........)))).))))).))).. ( -36.10) >consensus UACCGAUAGCUUAUAUCGGCGGCUGUGGGUA______________ACAGGGUUGUU____GCCUAUCGAUAGACAACAGCUG_GAGCGAUAGAUGGCGCUAGUACCAAGUU_ (((((((((((.........)))))))))))..................((((((((.(((((....))))).))))))))...((((........))))............ (-18.35 = -20.85 + 2.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:10 2006