| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,670,143 – 18,670,244 |
| Length | 101 |
| Max. P | 0.667736 |

| Location | 18,670,143 – 18,670,244 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -16.16 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18670143 101 + 22224390 UA------------UGGCUAUGUUUACAUCUUUCUCACUUCUUCGUCUUCUAUU-UUUUGUUUUGCAUUUUAAUGAAUGAACGCCUUUUGCGAUUUUACAAUAGCCAUUUAUAU------ .(------------(((((((((....(((........................-....((((..(((....)))...))))((.....)))))...)).))))))))......------ ( -15.10) >DroSec_CAF1 15090 101 + 1 UA------------UGGCUAUGUUUACAUCUUUCUCACUUCUUCGUCUUCUAUU-UUUUGUUUUGCAUUUUAAUGAAUGAACGCCUUUUGCGAUUUUACAAUAGCCAUUUAUAU------ .(------------(((((((((....(((........................-....((((..(((....)))...))))((.....)))))...)).))))))))......------ ( -15.10) >DroSim_CAF1 13215 101 + 1 UA------------UGGCUAUGUUUACAUCUUUCUCACUUCUUCGUCUUCUAUU-UUUUGUUUUGCAUUUUAAUGAAUGAACGCCUUUUGCGAUUUUACAAUAGCCAUUUAUAU------ .(------------(((((((((....(((........................-....((((..(((....)))...))))((.....)))))...)).))))))))......------ ( -15.10) >DroEre_CAF1 15281 108 + 1 UAU-------UAUAUGGCUAUGUUUACAUCUGCCUCACUUCUUCGUCUUAUUUUGUUUUGUUUUGCAUUUUAAUGAAUGAACGACUGUUGCGAUUUUACAAUAGCCAUUUAUUUA----- ...-------((.((((((((((....(((.((..((.((.(((((.(((((.(((........)))....)))))))))).)).))..)))))...)).)))))))).))....----- ( -20.90) >DroYak_CAF1 15192 119 + 1 UACAUAUAUGUAUGUAGCUAUGUUUACAUCUUCCCCACUUCCUCGUCUUCUAUU-UUUUGUUUUGCAUUUUAAUGAAUGAACGAUUGUUGCGAUUUUACAAUAGCCAUUUAUAUGUUGUU .(((.(((((((.((.(((((((....(((......((......))........-..((((((..(((....)))...)))))).......)))...)).))))).)).)))))))))). ( -14.60) >consensus UA____________UGGCUAUGUUUACAUCUUUCUCACUUCUUCGUCUUCUAUU_UUUUGUUUUGCAUUUUAAUGAAUGAACGCCUUUUGCGAUUUUACAAUAGCCAUUUAUAU______ ..............(((((((((....(((.............................((((..(((....)))...))))((.....)))))...)).)))))))............. (-11.04 = -11.64 + 0.60)

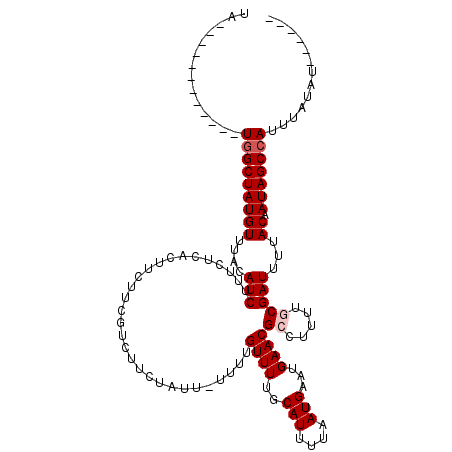

| Location | 18,670,143 – 18,670,244 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18670143 101 - 22224390 ------AUAUAAAUGGCUAUUGUAAAAUCGCAAAAGGCGUUCAUUCAUUAAAAUGCAAAACAAAA-AAUAGAAGACGAAGAAGUGAGAAAGAUGUAAACAUAGCCA------------UA ------......((((((((.((...(((((.....)).((((((..((......(.........-....)......))..))))))...)))....)))))))))------------). ( -15.32) >DroSec_CAF1 15090 101 - 1 ------AUAUAAAUGGCUAUUGUAAAAUCGCAAAAGGCGUUCAUUCAUUAAAAUGCAAAACAAAA-AAUAGAAGACGAAGAAGUGAGAAAGAUGUAAACAUAGCCA------------UA ------......((((((((.((...(((((.....)).((((((..((......(.........-....)......))..))))))...)))....)))))))))------------). ( -15.32) >DroSim_CAF1 13215 101 - 1 ------AUAUAAAUGGCUAUUGUAAAAUCGCAAAAGGCGUUCAUUCAUUAAAAUGCAAAACAAAA-AAUAGAAGACGAAGAAGUGAGAAAGAUGUAAACAUAGCCA------------UA ------......((((((((.((...(((((.....)).((((((..((......(.........-....)......))..))))))...)))....)))))))))------------). ( -15.32) >DroEre_CAF1 15281 108 - 1 -----UAAAUAAAUGGCUAUUGUAAAAUCGCAACAGUCGUUCAUUCAUUAAAAUGCAAAACAAAACAAAAUAAGACGAAGAAGUGAGGCAGAUGUAAACAUAGCCAUAUA-------AUA -----.......((((((((.((...(((((.((..(((((((((......))))..................)))))....))...)).)))....))))))))))...-------... ( -17.46) >DroYak_CAF1 15192 119 - 1 AACAACAUAUAAAUGGCUAUUGUAAAAUCGCAACAAUCGUUCAUUCAUUAAAAUGCAAAACAAAA-AAUAGAAGACGAGGAAGUGGGGAAGAUGUAAACAUAGCUACAUACAUAUAUGUA ....(((((((..(((((((.((...(((.(..((.(((((..(((...................-....)))))))).....))..)..)))....)))))))))......))))))). ( -16.60) >consensus ______AUAUAAAUGGCUAUUGUAAAAUCGCAAAAGGCGUUCAUUCAUUAAAAUGCAAAACAAAA_AAUAGAAGACGAAGAAGUGAGAAAGAUGUAAACAUAGCCA____________UA .............(((((((.((...((((((.....................)))..................((......))......)))....))))))))).............. (-11.42 = -11.26 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:06 2006