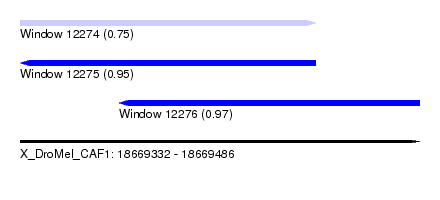
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,669,332 – 18,669,486 |
| Length | 154 |
| Max. P | 0.965629 |

| Location | 18,669,332 – 18,669,446 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.92 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18669332 114 + 22224390 GAAAACAAUGAUAGCUGAAUUCC-GUGUGCGUGUGUGUG--UGUUGAUUGCAGCCCACAAAUAAACACUUGUGCACCCA--UUGCACCUAAA-ACGCAAGUGCAGAAACAUACACACAAA .............((........-....)).((((((((--((((..((((...................(((((....--.))))).....-((....)))))).)))))))))))).. ( -29.80) >DroSec_CAF1 14424 104 + 1 GAAAACAAUGAUAGCCGAUUUUC-GUG----------UG--UGUCGAUUGCAGCCCACAAAUAAACACUUGUGCACCCA--CUGCACCUAAA-ACACAAGUGCAGAAACACACACACAAA ((((((..........).)))))-(((----------((--(((...........(((((........)))))......--((((((.....-......))))))..))))))))..... ( -23.40) >DroSim_CAF1 12517 102 + 1 GAAA--CAUGAUAGCUGAUUUUC-GUG----------UG--UGUUGAUUACAGCCCACAAAUAAACACUUGUGCACCCA--CUGCACCUAAA-ACACAAGUGCAGAAACACACACACAAA ....--.................-(((----------((--((((..........(((((........)))))......--((((((.....-......)))))).)))))))))..... ( -24.70) >DroEre_CAF1 14629 115 + 1 GAAAACAGCGAUAGCUGCUUUCUUGUGUGCGUGAGUGUG--UGUUGAUUGCA-CCCACAAAUAAACACUUGCACACCCACGCUGCACCGAAACACGCAAGUGCAGAAACA--CACACAAA ((((.((((....)))).))))(((((((((((.(((((--((((..(((..-....)))...)))....)))))).))))((((((((.....))...)))))).....--))))))). ( -37.90) >DroYak_CAF1 14659 114 + 1 GAAAACAAUGAUAGCUGAUUUCU-GUGUGCGUGUGUGUGUCUGUUGAUUGCAACACACAAAUAAACAGUUGUACACCCA--CUGCACCUAAA-ACGCAAGUGCAGAAACA--CACACAAA ((((.((........)).))))(-(((((.((.(((((((.(((.....))))))))))....................--((((((.....-......))))))..)).--)))))).. ( -29.10) >consensus GAAAACAAUGAUAGCUGAUUUCC_GUGUGCGUG_GUGUG__UGUUGAUUGCAGCCCACAAAUAAACACUUGUGCACCCA__CUGCACCUAAA_ACGCAAGUGCAGAAACA_ACACACAAA ........................(((((......((((..(((.....)))...))))......................((((((............)))))).......)))))... (-11.64 = -12.92 + 1.28)

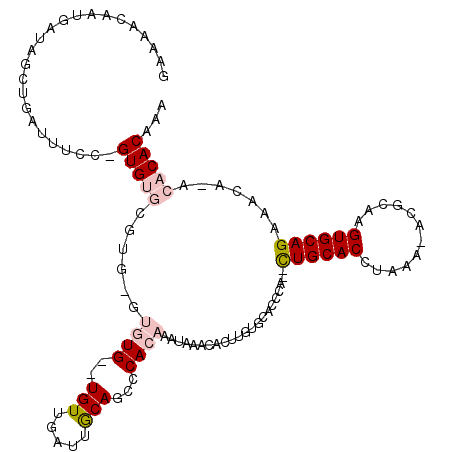
| Location | 18,669,332 – 18,669,446 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -19.06 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18669332 114 - 22224390 UUUGUGUGUAUGUUUCUGCACUUGCGU-UUUAGGUGCAA--UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCAACA--CACACACACGCACAC-GGAAUUCAGCUAUCAUUGUUUUC ((((((((..(((...((((((((...-..)))))))).--..(((.(((((((....))))))).)))..........--.....)))..)))))-))).................... ( -31.60) >DroSec_CAF1 14424 104 - 1 UUUGUGUGUGUGUUUCUGCACUUGUGU-UUUAGGUGCAG--UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCGACA--CA----------CAC-GAAAAUCGGCUAUCAUUGUUUUC (((((((((((....(((((((((...-..)))))))))--..(((.(((((((....))))))).))).......)))--))----------)))-))).................... ( -36.90) >DroSim_CAF1 12517 102 - 1 UUUGUGUGUGUGUUUCUGCACUUGUGU-UUUAGGUGCAG--UGGGUGCACAAGUGUUUAUUUGUGGGCUGUAAUCAACA--CA----------CAC-GAAAAUCAGCUAUCAUG--UUUC (((((((((((....(((((((((...-..)))))))))--..(((.(((((((....))))))).))).......)))--))----------)))-)))..............--.... ( -36.80) >DroEre_CAF1 14629 115 - 1 UUUGUGUG--UGUUUCUGCACUUGCGUGUUUCGGUGCAGCGUGGGUGUGCAAGUGUUUAUUUGUGGG-UGCAAUCAACA--CACACUCACGCACACAAGAAAGCAGCUAUCGCUGUUUUC .(((((((--(....(((((((..........))))))).((((((((....(((((...((((...-.))))..))))--)))))))))))))))))(((((((((....))))))))) ( -46.80) >DroYak_CAF1 14659 114 - 1 UUUGUGUG--UGUUUCUGCACUUGCGU-UUUAGGUGCAG--UGGGUGUACAACUGUUUAUUUGUGUGUUGCAAUCAACAGACACACACACGCACAC-AGAAAUCAGCUAUCAUUGUUUUC ((((((((--(((...((((((((...-..))))))))(--((.((((....(((((...((((.....))))..))))))))).)))))))))))-))).................... ( -35.90) >consensus UUUGUGUGU_UGUUUCUGCACUUGCGU_UUUAGGUGCAG__UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCAACA__CACAC_CACGCACAC_GGAAAUCAGCUAUCAUUGUUUUC ...(((((..((((.(((((((((......)))))))))....(((.(((((((....))))))).)))......))))..))))).................................. (-19.06 = -20.38 + 1.32)

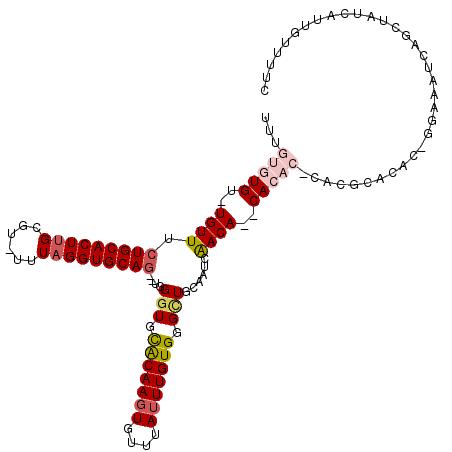
| Location | 18,669,370 – 18,669,486 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -30.08 |
| Energy contribution | -31.56 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18669370 116 - 22224390 UUUAAGCUCGCCUUGUAACGGUACUCCUCGGGAGUUUCAAUUUGUGUGUAUGUUUCUGCACUUGCGU-UUUAGGUGCAA--UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCAACA- ....(((((((.....(((((.(((((...))))).))......(((((((.(...((((((((...-..)))))))).--.).)))))))...))).....)))))))..........- ( -34.60) >DroSec_CAF1 14452 116 - 1 UUUAAGCUCGCCUUGUAACGGGACUCCACGGGAGUUUCAAUUUGUGUGUGUGUUUCUGCACUUGUGU-UUUAGGUGCAG--UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCGACA- ....(((((((.....((((..(((((...)))))..).(((((((..(.(....(((((((((...-..)))))))))--.).)..)))))))))).....)))))))..........- ( -42.50) >DroSim_CAF1 12543 116 - 1 UUUAAGCUCGCCUUGUAACGGGACUCCACGGGAGUUUCAAUUUGUGUGUGUGUUUCUGCACUUGUGU-UUUAGGUGCAG--UGGGUGCACAAGUGUUUAUUUGUGGGCUGUAAUCAACA- ....(((((((.....((((..(((((...)))))..).(((((((..(.(....(((((((((...-..)))))))))--.).)..)))))))))).....)))))))..........- ( -42.50) >DroEre_CAF1 14668 116 - 1 UUUAAGCUCGCCUUGUAACGGUACUCCUCUGCAGCUUCAAUUUGUGUG--UGUUUCUGCACUUGCGUGUUUCGGUGCAGCGUGGGUGUGCAAGUGUUUAUUUGUGGG-UGCAAUCAACA- .....((((((.....(((.((((.((((((((.(...(((.((((.(--(((....)))).)))).)))..).)))))...))).))))....))).....)))))-)..........- ( -29.40) >DroYak_CAF1 14698 115 - 1 UUUAAGCUCGCCUUGUUACGGGACUCCUUGGGAGUUUCAAUUUGUGUG--UGUUUCUGCACUUGCGU-UUUAGGUGCAG--UGGGUGUACAACUGUUUAUUUGUGUGUUGCAAUCAACAG .....(..(((((......(..(((((...)))))..)..........--.....(((((((((...-..)))))))))--.)))))..)..(((((...((((.....))))..))))) ( -32.30) >consensus UUUAAGCUCGCCUUGUAACGGGACUCCUCGGGAGUUUCAAUUUGUGUGU_UGUUUCUGCACUUGCGU_UUUAGGUGCAG__UGGGUGCACAAGUGUUUAUUUGUGGGCUGCAAUCAACA_ ....(((((((.....(((((((((((...)))))))).(((((((((.......(((((((((......)))))))))......)))))))))))).....)))))))........... (-30.08 = -31.56 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:04 2006