
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,665,590 – 18,665,750 |
| Length | 160 |
| Max. P | 0.748149 |

| Location | 18,665,590 – 18,665,710 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.18 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -28.94 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
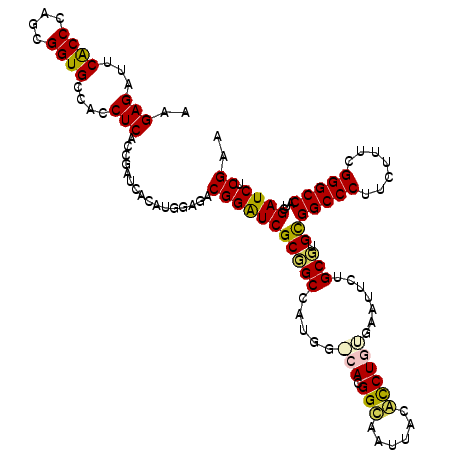
>X_DroMel_CAF1 18665590 120 + 22224390 AAGAGAUUCACCCAGCGGUGCCACCUCACCGAUCACAUAAAGACGGAUCGCGGCCAUGGACACGGCACUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUCCGGGCCAUCGAUCUCGAA ..(((((((((.((((.((((((...(..(((((.(........))))))..)...))).))).))..((((....))))....)).))).(((((.......)))))...))))))... ( -38.20) >DroSec_CAF1 10640 114 + 1 AAGAGAUUCACCCAGCGGUGCCACCUCACCGAUCACAUGGGGACGGAUCGCUGCC------ACGGUAAUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUUCGGGCCAUCGAUCUCGAA ..(((((((((.(((((((.((.(((((.........)))))..))))))))).(------(((((......))).)))........))).(((((.......)))))...))))))... ( -39.30) >DroSim_CAF1 8747 114 + 1 AAGAGAUUCACCCAGCGGUGCCACCUCACCGAUCACAUGGAGACGGGUCGCUGCC------ACGGUAAUUACACCUGUGAAUUAUGCGUGCGGCCCUUCUCUCGGGCCAUCGAUCUCGAA ..((((((..(((..(((((......))))).......(((((.(((((((((((------(((((......))).)))......))).))))))).))))).))).....))))))... ( -43.30) >DroEre_CAF1 10950 120 + 1 AAGAGAUUCGCCCAGCGGUGCCACCUCAUCCAGCACUUGGCCACGGAUCGCGGCCAUGGGCACGGCAAGUACGUCUGCCAACUCUGCCUGUGGCCCUUCUUUCGGGCCAUCGAUCUCGAA ..(((((((((...)))((((...........)))).(((((.((((....(((((((((((.((((........)))).....))))))))))).....)))))))))..))))))... ( -48.20) >DroYak_CAF1 10860 120 + 1 AAGAGAUUCGCCCAGCGGUGCCACCUCACCCAGCACAUGGCCACGGAUCGCGGCCAUGGGCAAGGCAACUUCGUCUGCCAACUCUGCCUGUGGCCCUUCUUCCGGGCCAUCGAUCUCGAA ..(((((.((....((((((......))))..))..((((((.((((....(((((((((((.((((........)))).....))))))))))).....)))))))))))))))))... ( -52.90) >consensus AAGAGAUUCACCCAGCGGUGCCACCUCACCGAUCACAUGGAGACGGAUCGCGGCCAUGG_CACGGCAAUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUUCGGGCCAUCGAUCUCGAA ..(((...((((....))))....)))................(((((((((((.....(((.(((......)))))).......))).))(((((.......)))))...)))).)).. (-28.94 = -28.46 + -0.48)


| Location | 18,665,630 – 18,665,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -44.86 |
| Consensus MFE | -31.56 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
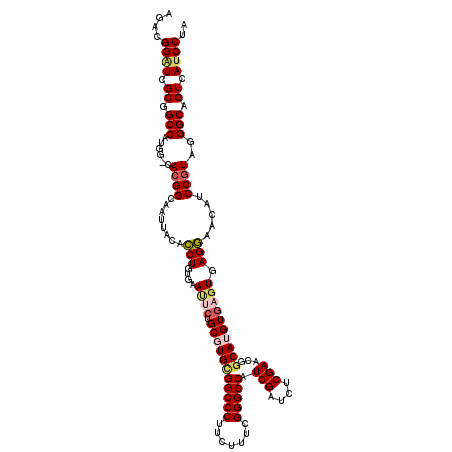
>X_DroMel_CAF1 18665630 120 + 22224390 AGACGGAUCGCGGCCAUGGACACGGCACUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUCCGGGCCAUCGAUCUCGAACGGCAUGUGUGUGAGGAACAUCCGUAGGGCAGUCAACCUA ..((((((....(((........))).(((((((...........(((((((((((.......))))).(((....)))...)))))))))))))....))))))(((........))). ( -42.00) >DroSec_CAF1 10680 114 + 1 GGACGGAUCGCUGCC------ACGGUAAUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUUCGGGCCAUCGAUCUCGAACGGCAUGUGAGUGAGGCACAUCCGUAGGGCAGUCAUCCUA ....((((.((((((------..(((......)))((((.((((.(((((((((((.......))))).(((....)))...))))))))))....)))).......)))))).)))).. ( -46.40) >DroSim_CAF1 8787 114 + 1 AGACGGGUCGCUGCC------ACGGUAAUUACACCUGUGAAUUAUGCGUGCGGCCCUUCUCUCGGGCCAUCGAUCUCGAACGGCAUGUGAGUGAGGAACAUCCGUAGGGCAGUCAUCCUA ....((((.((((((------((((...((((....)))).(((((((((((((((.......))))).(((....)))...))))))..)))).......))))..)))))).)))).. ( -42.50) >DroEre_CAF1 10990 120 + 1 CCACGGAUCGCGGCCAUGGGCACGGCAAGUACGUCUGCCAACUCUGCCUGUGGCCCUUCUUUCGGGCCAUCGAUCUCGAACGCCAUGUGAUUAAGAAGCAUCCGUAGGGCAGUCAUCCUA ((((((((.(((((((((((((.((((........)))).....))))))))))).(((((((.(((..(((....)))..)))....))..))))))))))))).))............ ( -45.80) >DroYak_CAF1 10900 120 + 1 CCACGGAUCGCGGCCAUGGGCAAGGCAACUUCGUCUGCCAACUCUGCCUGUGGCCCUUCUUCCGGGCCAUCGAUCUCGAACGGCAUGUGUGUAAGAAGCAUCCGUGAGGCAGUCAUCCUA .(((((((.(((((((((((((.((((........)))).....))))))))))).((((((((.(((.(((....)))..)))...)).).))))))))))))))(((.......))). ( -47.60) >consensus AGACGGAUCGCGGCCAUGG_CACGGCAAUUACACCUGUGAAUUCUGCGUGCGGCCCUUCUUUCGGGCCAUCGAUCUCGAACGGCAUGUGAGUGAGGAACAUCCGUAGGGCAGUCAUCCUA ....((((.((.(((......((((........(((....((((.(((((((((((.......))))).(((....)))...)))))))))).))).....))))..))).)).)))).. (-31.56 = -32.52 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:58 2006