
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,654,928 – 18,655,059 |
| Length | 131 |
| Max. P | 0.909865 |

| Location | 18,654,928 – 18,655,040 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.83 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
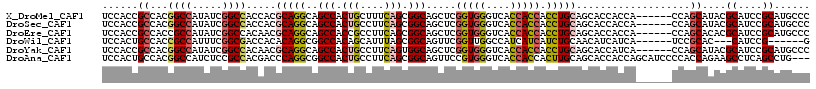
>X_DroMel_CAF1 18654928 112 + 22224390 UCCACCGCCACGGCCAUAUCGGCCACCACGCAGGCAGCCACUGCUUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCAUACGCAUCCGCAUGCCC ......((...((((.....)))).....(((((..(((.(((....))).))).....(((((....))))).))))).)).......------...((((.((....)).)))).. ( -40.70) >DroSec_CAF1 43 112 + 1 UCCACCGCCACGGCCAUAUCGGCCACCACGCAGGCAGCCACUGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCAUACGCAUCCGCAUGCCC ......((...((((.....)))).....(((((..(((.(((....))).))).....(((((....))))).))))).)).......------...((((.((....)).)))).. ( -40.70) >DroEre_CAF1 482 112 + 1 UCCACCGCCACCGCCAUAUCGGCCACAACGCAGGCAGCCACCGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCACACGCAUCCGCAUGCCC ......((....(((.....)))......(((((.(((..((((.....))))..))).(((((....))))).))))).)).......------...(((..((....))..))).. ( -35.70) >DroWil_CAF1 397 103 + 1 UCCACUGCCACCGCCAUUUCGGCGACCACACAGGCGGCCACAGCAUUUAGCGGCAGUUCGGUUGGCCAUCAUCAUCUGCAACAUCAUCA------UCCGCAC---CAUCCG------G ...((((((.(((((.....(....)......))))).....((.....)))))))).(((.(((...........(((..........------...))))---)).)))------. ( -26.32) >DroYak_CAF1 343 112 + 1 UCCACCGCCACGGCCAUAUCGGCCACAACGCAGGCAGCCACUGCCUUCAGUGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCAUCA------CCAGCAUACGCAUCCGCAUGCCC ......((...((((.....)))).....(((((..(((((((....))))))).....(((((....))))).))))).)).......------...((((.((....)).)))).. ( -44.60) >DroAna_CAF1 5848 115 + 1 UCCACUGCCACGGCCAUCUCCGCCACGACCCAGGCGGCCACUGCCUUCAGCGGCAGUUCCGUGGGUCACCACCACUUGCAGCACCACCAGCAUCCCCACCAGAAGCCUCAGCCUG--- ....((((...(((.......)))..(((((..((((..((((((......)))))).)))))))))..........))))........((.((.......)).)).........--- ( -33.40) >consensus UCCACCGCCACGGCCAUAUCGGCCACCACGCAGGCAGCCACUGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA______CCAGCAUACGCAUCCGCAUGCCC ......((...((((.....)))).....(((((..(((.(((....))).))).....(((((....))))).)))))...................))....((....))...... (-26.20 = -26.83 + 0.63)

| Location | 18,654,928 – 18,655,040 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -52.03 |
| Consensus MFE | -36.90 |
| Energy contribution | -39.37 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18654928 112 - 22224390 GGGCAUGCGGAUGCGUAUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAAGCAGUGGCUGCCUGCGUGGUGGCCGAUAUGGCCGUGGCGGUGGA .((((((((....)))))))).(------..(.....)..).(((((.(....))))))...(..((((((...((..((((..((.....))..)))).....))..))))))..). ( -53.90) >DroSec_CAF1 43 112 - 1 GGGCAUGCGGAUGCGUAUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCAGUGGCUGCCUGCGUGGUGGCCGAUAUGGCCGUGGCGGUGGA .((((((((....)))))))).(------..(.....)..).(((((.(....))))))...(..(((((...(((..((((..((.....))..)))).....)))..)))))..). ( -57.50) >DroEre_CAF1 482 112 - 1 GGGCAUGCGGAUGCGUGUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCGGUGGCUGCCUGCGUUGUGGCCGAUAUGGCGGUGGCGGUGGA .((((((((....))))))))..------.......((((((((((((((....))))).(((..((((.....))))..)))))))))((..(.(((.....))).)..)))))... ( -55.80) >DroWil_CAF1 397 103 - 1 C------CGGAUG---GUGCGGA------UGAUGAUGUUGCAGAUGAUGAUGGCCAACCGAACUGCCGCUAAAUGCUGUGGCCGCCUGUGUGGUCGCCGAAAUGGCGGUGGCAGUGGA .------(((.((---((.((..------...((......)).....))...)))).))).(((((((((.........((((((....))))))(((.....))))))))))))... ( -37.10) >DroYak_CAF1 343 112 - 1 GGGCAUGCGGAUGCGUAUGCUGG------UGAUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCACUGAAGGCAGUGGCUGCCUGCGUUGUGGCCGAUAUGGCCGUGGCGGUGGA .((((((((....))))))))..------.......((((((((((((((....))))).....(((((((....))))))).))))))((..(((((.....)))))..)))))... ( -56.80) >DroAna_CAF1 5848 115 - 1 ---CAGGCUGAGGCUUCUGGUGGGGAUGCUGGUGGUGCUGCAAGUGGUGGUGACCCACGGAACUGCCGCUGAAGGCAGUGGCCGCCUGGGUCGUGGCGGAGAUGGCCGUGGCAGUGGA ---...((((.(((((((.((.(.(((.(.(((((((((((.(((((..((..(....)..))..)))))....))))).)))))).).))).).)).).)).))))....))))... ( -51.10) >consensus GGGCAUGCGGAUGCGUAUGCUGG______UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCAGUGGCUGCCUGCGUGGUGGCCGAUAUGGCCGUGGCGGUGGA .((((((((....))))))))...............((((((((((((((....))))).....(((((((....))))))).))))))((..(((((.....)))))..)))))... (-36.90 = -39.37 + 2.46)


| Location | 18,654,968 – 18,655,059 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.70 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
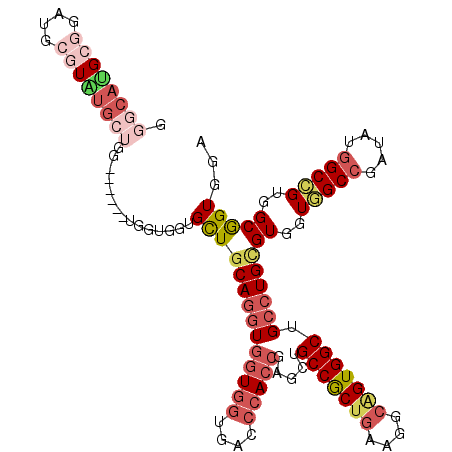
>X_DroMel_CAF1 18654968 91 + 22224390 CUGCUUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCAUACGCAUCCGCAUGCCC---CACAGCAGUCACAGUCACA------------------- (((((....((.((((...(((((....)))))..)))).)).......------...((((.((....)).))))..---...)))))...........------------------- ( -27.50) >DroSec_CAF1 83 90 + 1 CUGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCAUACGCAUCCGCAUGCCC---CACAGCAGUCACAGUCAC-------------------- ((((.....((.((((...(((((....)))))..)))).)).......------...((((.((....)).))))..---....))))..........-------------------- ( -26.50) >DroEre_CAF1 522 110 + 1 CCGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA------CCAGCACACGCAUCCGCAUGCCC---CACAGCAGUCGCAGUCACAGUCCCAGUCACUGACACAG ......((((.(((.....(((((....)))))..((((.((.......------.........((....)).(((..---....))))).))))...........))).))))..... ( -28.40) >DroYak_CAF1 383 104 + 1 CUGCCUUCAGUGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCAUCA------CCAGCAUACGCAUCCGCAUGCCC---CACAGCAGUCC------CAGUCCCAGUCACAGCCACAU .........(((((.....(((((....)))))..((((.((.......------...))....((((....))))..---....))))...------..............))))).. ( -28.20) >DroAna_CAF1 5888 86 + 1 CUGCCUUCAGCGGCAGUUCCGUGGGUCACCACCACUUGCAGCACCACCAGCAUCCCCACCAGAAGCCUCAGCCUG--------CGGCAGUGACG------------------------- (((((......)))))....((((....))))((((.((.(((......((.((.......)).)).......))--------).))))))...------------------------- ( -23.42) >DroPer_CAF1 8535 95 + 1 CCGCAUUUAGCGGCAGUUCCGUGGGCCACCAUCACUUGCAGCACCACCA---------------GCAUCCGCACGCCCAUUCACAGCCCGCAACGCACC---------AACAACAACUG ((((.....))))(((((..((((((..........(((.((.......---------------))....)))............))))))...(....---------..)...))))) ( -17.95) >consensus CUGCCUUCAGCGGCAGCUCGGUGGGUCACCACCACCUGCAGCACCACCA______CCAGCAUACGCAUCCGCAUGCCC___CACAGCAGUCACAGUCAC____________________ ((((.....((.((((...(((((....)))))..)))).))......................((....)).............)))).............................. (-19.23 = -19.57 + 0.34)


| Location | 18,654,968 – 18,655,059 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 68.70 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -26.34 |
| Energy contribution | -28.90 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18654968 91 - 22224390 -------------------UGUGACUGUGACUGCUGUG---GGGCAUGCGGAUGCGUAUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAAGCAG -------------------...........(((((...---.((((((((....))))))))((------(((..((......(((((.(....))))))..))..)))))...))))) ( -41.10) >DroSec_CAF1 83 90 - 1 --------------------GUGACUGUGACUGCUGUG---GGGCAUGCGGAUGCGUAUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCAG --------------------..........(((((...---.((((((((....))))))))((------(((..((......(((((.(....))))))..))..)))))...))))) ( -41.10) >DroEre_CAF1 522 110 - 1 CUGUGUCAGUGACUGGGACUGUGACUGCGACUGCUGUG---GGGCAUGCGGAUGCGUGUGCUGG------UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCGG ...((.((((.((.......)).)))))).(((((...---.((((((((....))))))))((------(((..((......(((((.(....))))))..))..)))))...))))) ( -46.00) >DroYak_CAF1 383 104 - 1 AUGUGGCUGUGACUGGGACUG------GGACUGCUGUG---GGGCAUGCGGAUGCGUAUGCUGG------UGAUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCACUGAAGGCAG .((..((.((..(.......)------..)).(((((.---(((((((((....))))))))..------).)))))))..))(((((.(....))))))...(((((......))))) ( -39.50) >DroAna_CAF1 5888 86 - 1 -------------------------CGUCACUGCCG--------CAGGCUGAGGCUUCUGGUGGGGAUGCUGGUGGUGCUGCAAGUGGUGGUGACCCACGGAACUGCCGCUGAAGGCAG -------------------------.(((((..(((--------(..((.(..(((.(..((......))..).))).).))..))))..)))))........(((((......))))) ( -36.40) >DroPer_CAF1 8535 95 - 1 CAGUUGUUGUU---------GGUGCGUUGCGGGCUGUGAAUGGGCGUGCGGAUGC---------------UGGUGGUGCUGCAAGUGAUGGUGGCCCACGGAACUGCCGCUAAAUGCGG (((((......---------...((((((((.(((.......))).))).)))))---------------..((((.((..(.(....).)..))))))..)))))((((.....)))) ( -33.90) >consensus ____________________GUGACUGUGACUGCUGUG___GGGCAUGCGGAUGCGUAUGCUGG______UGGUGGUGCUGCAGGUGGUGGUGACCCACCGAGCUGCCGCUGAAGGCAG ..............................(((((.......((((((((....)))))))).........(((((((((...(((((.(....)))))).))).))))))...))))) (-26.34 = -28.90 + 2.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:50 2006