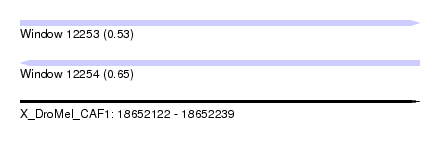
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,652,122 – 18,652,239 |
| Length | 117 |
| Max. P | 0.652438 |

| Location | 18,652,122 – 18,652,239 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18652122 117 + 22224390 CUAGAAGCGUUCACUCUGCACAUACAUAAGGUGGGCACCUAAUGCUCUGGUGCCCAUGAGCAUGAGCCUAGAGCUCCAGAGCUCCAGAGCUCCGCUUGAUUGACAAAUGAACAAAUC ........(((((...((((..........(((((((((.........)))))))))((((..((((((.(((((....))))).)).)))).))))...)).))..)))))..... ( -41.60) >DroSec_CAF1 11067 104 + 1 CUAGAAGCGACCACUCUGCACAUACAUAAGGUGGGCACCUAAUGCUCUG---CC--CGAGCAUGAGCCUAGAGCUCC--------AGAGCUCCGCUUGAUUGACAAAUGAACAAAUC ....(((((......................(((((.....((((((..---..--.))))))..)))))(((((..--------..)))))))))).................... ( -26.70) >DroSim_CAF1 11174 104 + 1 CUAGAAGCGGCCACUCGGCACACACAUAAGGUGGGCACCUAAUGCUCUG---CC--CGAGCAUGAGCCUAGAGCUCC--------AGAGCUCCGCUUGAUUGACAAAUGAACAAAUC ....((((((((....)))............(((((.....((((((..---..--.))))))..)))))(((((..--------..)))))))))).................... ( -30.80) >consensus CUAGAAGCGACCACUCUGCACAUACAUAAGGUGGGCACCUAAUGCUCUG___CC__CGAGCAUGAGCCUAGAGCUCC________AGAGCUCCGCUUGAUUGACAAAUGAACAAAUC ....(((((......................(((((.....((((((..........))))))..)))))((((((..........))))))))))).................... (-27.20 = -27.20 + -0.00)

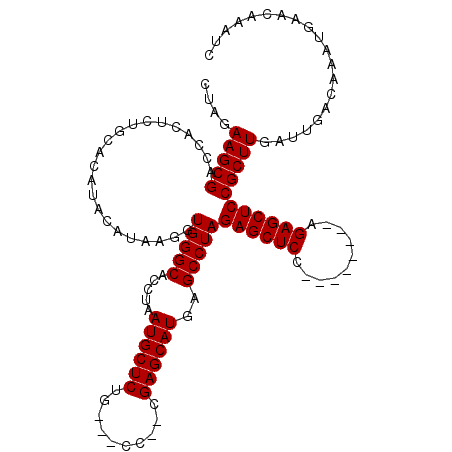
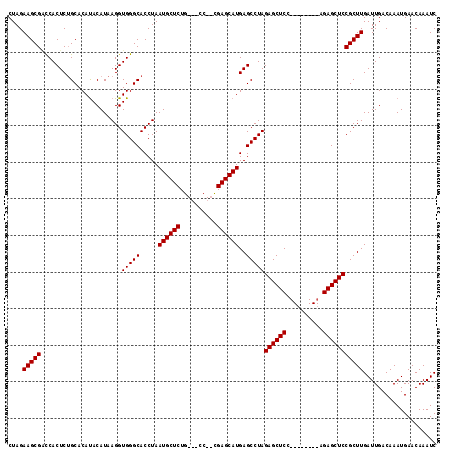
| Location | 18,652,122 – 18,652,239 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -31.37 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18652122 117 - 22224390 GAUUUGUUCAUUUGUCAAUCAAGCGGAGCUCUGGAGCUCUGGAGCUCUAGGCUCAUGCUCAUGGGCACCAGAGCAUUAGGUGCCCACCUUAUGUAUGUGCAGAGUGAACGCUUCUAG ((..(((((((((..((..((((((((((.((((((((....)))))))))))).))))..((((((((.........)))))))).....))....))..)))))))))..))... ( -49.30) >DroSec_CAF1 11067 104 - 1 GAUUUGUUCAUUUGUCAAUCAAGCGGAGCUCU--------GGAGCUCUAGGCUCAUGCUCG--GG---CAGAGCAUUAGGUGCCCACCUUAUGUAUGUGCAGAGUGGUCGCUUCUAG ((((..(......)..))))(((((((((.((--------((....))))))))..((((.--.(---((.(((((.((((....)))).)))).).))).))))...))))).... ( -33.30) >DroSim_CAF1 11174 104 - 1 GAUUUGUUCAUUUGUCAAUCAAGCGGAGCUCU--------GGAGCUCUAGGCUCAUGCUCG--GG---CAGAGCAUUAGGUGCCCACCUUAUGUGUGUGCCGAGUGGCCGCUUCUAG ((((..(......)..))))((((((((((..--------..)))))).((((...(((((--((---((..((((.((((....)))).)))).))).)))))))))))))).... ( -38.40) >consensus GAUUUGUUCAUUUGUCAAUCAAGCGGAGCUCU________GGAGCUCUAGGCUCAUGCUCG__GG___CAGAGCAUUAGGUGCCCACCUUAUGUAUGUGCAGAGUGGACGCUUCUAG ((((..(......)..))))((((((((((((((....)))))))))...((((((((((..........)))))).((((....))))............))))...))))).... (-31.37 = -31.70 + 0.33)

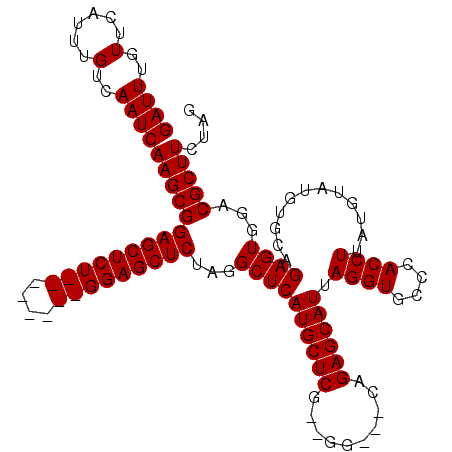

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:44 2006