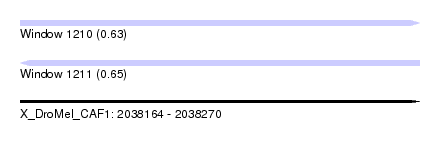
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,038,164 – 2,038,270 |
| Length | 106 |
| Max. P | 0.651372 |

| Location | 2,038,164 – 2,038,270 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
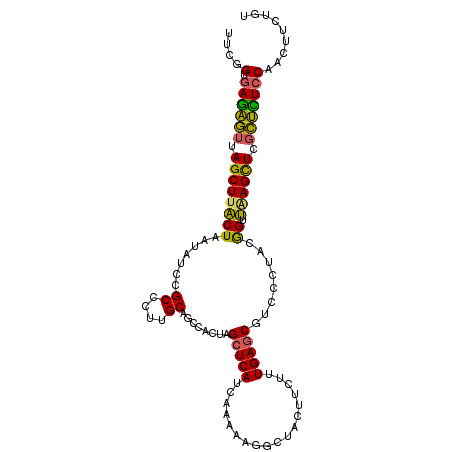
>X_DroMel_CAF1 2038164 106 + 22224390 ACAGAAGUUGGAGAGCGAGCUUAACCGUAAGGACGCUCAAAGAAGUAGCCUUUUUGAUGAGCUAGUGGCUGCAAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA ..........(((((..((((((.((....))..(((((((((((....))))))..)))))(((((.((((....)))).))))).))))))..)))))...... ( -33.90) >DroSec_CAF1 4858 106 + 1 ACAGAAGUUGGAGAGCGAGCUUAACCGUAAGGAAGCUCAAAGAAGUGGCCUUUUUGAUGAGCUAGUGGCUGCGAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA ..........(((((..((((((.((....))..(((((((((((....))))))..)))))(((((.((((....)))).))))).))))))..)))))...... ( -34.00) >DroSim_CAF1 4843 106 + 1 ACAGAAGUUGGAGAGCGAGCUUAACCGUAAGGACGCUCAAAGAAGUGGCCUUUUUGAUGAGCUAGUGGCUGCGAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA ..........(((((..((((((.((....))..(((((((((((....))))))..)))))(((((.((((....)))).))))).))))))..)))))...... ( -34.10) >DroEre_CAF1 4880 106 + 1 ACAGAAGUUGGAAAGCGAACUUAACUAUCGCGUCGCUCAGAGAAGCAGCCUGUUUGAUGAGCUAGUGGCUGCAAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA ...((((((((...((((.........)))).((((((......((((((.((((...))))....))))))..))))))...........)))))).))...... ( -28.10) >DroYak_CAF1 4850 106 + 1 ACAGAAGUUGGAAAGCGAGCUCAACUAUGGAGUCGCUCAAAGAAGCAGCCUGUUUGAUGAGCUGGUGGCUGCAAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA .......((((..((..((((..((((.....((((((......((((((.((((...))))....))))))..))))))....)))).))))....))..)))). ( -28.60) >DroAna_CAF1 5215 106 + 1 ACAGAACCUGGAACUGGAGCUGGGCCUGGGCAAGGAUCAGCGUCCCCAGCUGUUCGAUGAACUGGUGGCCGCCAGGGCCGACAUAAGCAAGCUCAAGUUGGCGGAG ......(((((....(((((((..(((.....)))..)))).)))(((.(.((((...)))).).)))...)))))((((((...((....))...)))))).... ( -39.80) >consensus ACAGAAGUUGGAAAGCGAGCUUAACCGUAAGGACGCUCAAAGAAGCAGCCUGUUUGAUGAGCUAGUGGCUGCAAGGGCGGAUAUUAGUAAGCUAACUCUCACCGAA ..........(((((..((((((...........(((((..((((......))))..)))))(((((.((((....)))).))))).))))))..)))))...... (-23.13 = -23.75 + 0.62)

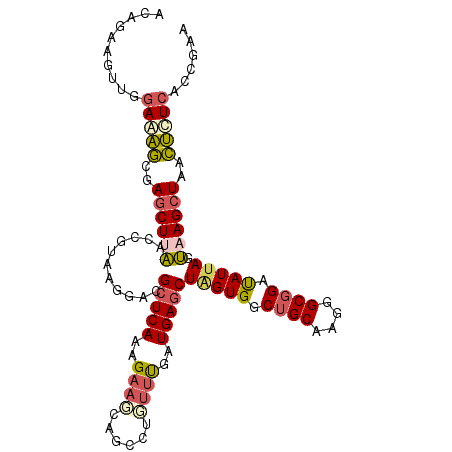
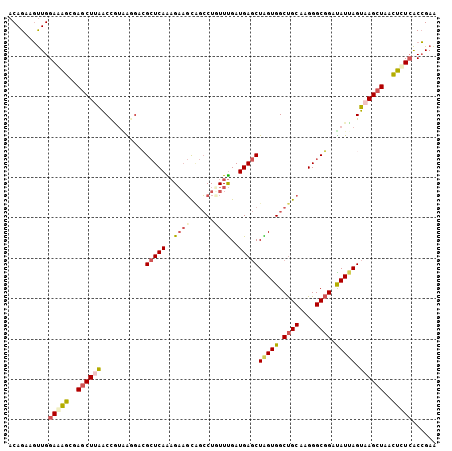
| Location | 2,038,164 – 2,038,270 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -20.51 |
| Energy contribution | -19.49 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2038164 106 - 22224390 UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUUGCAGCCACUAGCUCAUCAAAAAGGCUACUUCUUUGAGCGUCCUUACGGUUAAGCUCGCUCUCCAACUUCUGU ....(.((((((.(((((..........((....))((((....(((((.....(((....)))...)))))((....))))))))))).)))))))......... ( -26.50) >DroSec_CAF1 4858 106 - 1 UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUCGCAGCCACUAGCUCAUCAAAAAGGCCACUUCUUUGAGCUUCCUUACGGUUAAGCUCGCUCUCCAACUUCUGU ....(.((((((.(((((..........((....))((((...((((((.....(((....)))...)))))).......))))))))).)))))))......... ( -26.30) >DroSim_CAF1 4843 106 - 1 UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUCGCAGCCACUAGCUCAUCAAAAAGGCCACUUCUUUGAGCGUCCUUACGGUUAAGCUCGCUCUCCAACUUCUGU ....(.((((((.(((((..........((....))((((....(((((.....(((....)))...)))))((....))))))))))).)))))))......... ( -26.60) >DroEre_CAF1 4880 106 - 1 UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUUGCAGCCACUAGCUCAUCAAACAGGCUGCUUCUCUGAGCGACGCGAUAGUUAAGUUCGCUUUCCAACUUCUGU ....(.((((((.((((((....(((((((.(((((((((................))))))......))).).)).))))..)))))).)))))))......... ( -25.59) >DroYak_CAF1 4850 106 - 1 UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUUGCAGCCACCAGCUCAUCAAACAGGCUGCUUCUUUGAGCGACUCCAUAGUUGAGCUCGCUUUCCAACUUCUGU ....(.((((((.(((((((((.....(((.(..((((((................))))))......).)))......))).)))))).)))))))......... ( -27.69) >DroAna_CAF1 5215 106 - 1 CUCCGCCAACUUGAGCUUGCUUAUGUCGGCCCUGGCGGCCACCAGUUCAUCGAACAGCUGGGGACGCUGAUCCUUGCCCAGGCCCAGCUCCAGUUCCAGGUUCUGU ....(((.((.((((....)))).)).)))(((((.(((..((((((........))))))(((.((((..(((.....)))..))))))).))))))))...... ( -36.90) >consensus UUCGGUGAGAGUUAGCUUACUAAUAUCCGCCCUUGCAGCCACUAGCUCAUCAAAAAGGCUACUUCUUUGAGCGUCCCUACGGUUAAGCUCGCUCUCCAACUUCUGU ....(.((((((.((((((((.......((....))........(((((..................)))))........)).)))))).)))))))......... (-20.51 = -19.49 + -1.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:49 2006