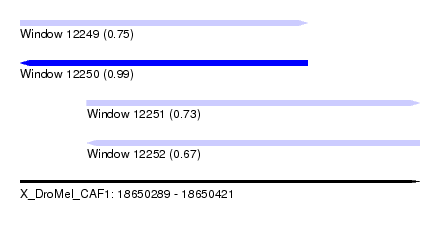
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,650,289 – 18,650,421 |
| Length | 132 |
| Max. P | 0.988811 |

| Location | 18,650,289 – 18,650,384 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -14.26 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18650289 95 + 22224390 CCUCAAAAGAACGUAAACAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCAAGAGCCCAGCAGC-------CAGCUGUAAU ................(((..(((((...((((((.....))))))....)))))(((((...(((.........))).))))).-------....)))... ( -21.60) >DroSec_CAF1 9147 95 + 1 CCUCAAAAGAACGUAAACAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC-------CAGCUGUAAU .(((.................(((((...((((((.....))))))....))))).(((.....)))....)))....((((...-------..)))).... ( -22.00) >DroSim_CAF1 9203 95 + 1 CCUCAAAAGAACGUAAACAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC-------CAGCUGUAAU .(((.................(((((...((((((.....))))))....))))).(((.....)))....)))....((((...-------..)))).... ( -22.00) >DroEre_CAF1 8452 75 + 1 CCUCAGAAGAACGUAAACAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAUGCCCCCUGCCUAAGGC---------------------------AGCUGUAAU ...(((...............(((((((.((((((.....))))))..)))))))(((((...)))---------------------------))))).... ( -22.60) >DroYak_CAF1 8524 102 + 1 CUUCAAAUGAACGUAAGCAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCCGCUUAAGGCAACUCGAGUGCACAGCACCCAGCACGCAGCUGUAAU ................((...(((((...((((((.....))))))....))))).((((..((....))..))))....))...((((.....)))).... ( -27.80) >consensus CCUCAAAAGAACGUAAACAAUGGGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC_______CAGCUGUAAU ................(((..(((((...((((((.....))))))....)))))..((.....))..............................)))... (-14.26 = -14.10 + -0.16)

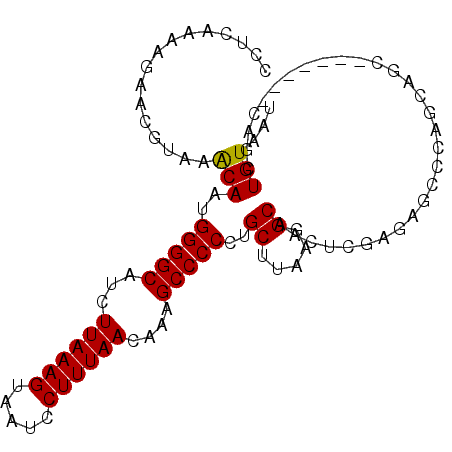
| Location | 18,650,289 – 18,650,384 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -23.94 |
| Energy contribution | -25.54 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18650289 95 - 22224390 AUUACAGCUG-------GCUGCUGGGCUCUUGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGUUUACGUUCUUUUGAGG ....((((..-------...))))..(((..(((..((..((((((((((((..(.((((((.....)))))).).))))).))))))).)).)))..))). ( -33.30) >DroSec_CAF1 9147 95 - 1 AUUACAGCUG-------GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGUUUACGUUCUUUUGAGG ....((((..-------...))))....((((((..((..((((((((((((..(.((((((.....)))))).).))))).))))))).))...)))))). ( -33.10) >DroSim_CAF1 9203 95 - 1 AUUACAGCUG-------GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGUUUACGUUCUUUUGAGG ....((((..-------...))))....((((((..((..((((((((((((..(.((((((.....)))))).).))))).))))))).))...)))))). ( -33.10) >DroEre_CAF1 8452 75 - 1 AUUACAGCU---------------------------GCCUUAGGCAGGGGGCAUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGUUUACGUUCUUCUGAGG ....(((((---------------------------(((...)))))((((((((.((((((.....))))))))))))))...............)))... ( -27.60) >DroYak_CAF1 8524 102 - 1 AUUACAGCUGCGUGCUGGGUGCUGUGCACUCGAGUUGCCUUAAGCGGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGCUUACGUUCAUUUGAAG ....((((.....)))).((((...))))((((((.((..((((((((((((..(.((((((.....)))))).).))))).))))))).))..)))))).. ( -35.30) >consensus AUUACAGCUG_______GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCCCAUUGUUUACGUUCUUUUGAGG ....((((............))))....((((((..((..((((((((((((..(.((((((.....)))))).).))))).))))))).))...)))))). (-23.94 = -25.54 + 1.60)



| Location | 18,650,311 – 18,650,421 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -25.89 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18650311 110 + 22224390 GGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCAAGAGCCCAGCAGC-------CAGCUGUAAUCGCAUCAACUGCGCCGUGCAACCA--GCACCA-UUGCACG ((((...((((((.....))))))....))))..(((....(((.......((...((((...-------..))))...))(((.....))))))((((.....--))))..-..))).. ( -30.20) >DroSec_CAF1 9169 112 + 1 GGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC-------CAGCUGUAAUCGCAUCAACUGCGCCGUGCAACCAGAGCACCA-UUGCACG ((((.((((..(((...((((.....((((.....))))))))..))).)))))))).((((.-------...))))...((((.....)))).(((((((...........-))))))) ( -30.20) >DroSim_CAF1 9225 112 + 1 GGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC-------CAGCUGUAAUCGCAUCAACUGCGCCGUGCAACCAGAGCGCCA-UUGCACG ((((.((((..(((...((((.....((((.....))))))))..))).)))))))).((((.-------...))))...((((.....)))).(((((((...........-))))))) ( -30.20) >DroYak_CAF1 8546 118 + 1 GGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCCGCUUAAGGCAACUCGAGUGCACAGCACCCAGCACGCAGCUGUAAUCACAUCAAUUGCGCCGUGCACAC--AGCACCAAAUGCACG ((((...((((((.....))))))....))))...(((..((....))..)((((((.((....(((.....)))(((((.......))))))).))))))..--.))............ ( -31.10) >consensus GGGCAUCUUAAAGUAAUCCUUUAACAAAGCCCCCUGCUUAAGGCAACUCGAGAGCCCAGCAGC_______CAGCUGUAAUCGCAUCAACUGCGCCGUGCAACCA_AGCACCA_UUGCACG ((((...((((((.....))))))....))))...((....(((.......((...((((............))))...))(((.....))))))((((.......)))).....))... (-25.89 = -26.20 + 0.31)

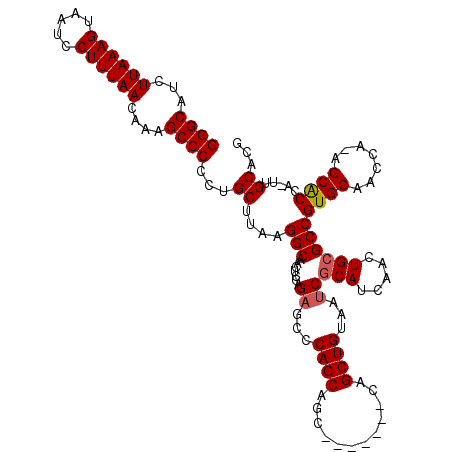
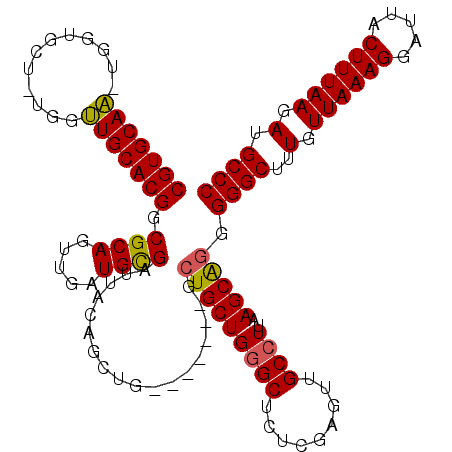
| Location | 18,650,311 – 18,650,421 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18650311 110 - 22224390 CGUGCAA-UGGUGC--UGGUUGCACGGCGCAGUUGAUGCGAUUACAGCUG-------GCUGCUGGGCUCUUGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCC (((((((-(.....--..))))))))..((((((...((.......)).)-------))))).((((((((((((..((((.((((((....)))))).))))...)).)))))).)))) ( -41.00) >DroSec_CAF1 9169 112 - 1 CGUGCAA-UGGUGCUCUGGUUGCACGGCGCAGUUGAUGCGAUUACAGCUG-------GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCC (((((((-(.(.....).))))))))...((((((.........))))))-------.(((((((((.........))))..))))).((((..(.((((((.....)))))).).)))) ( -39.60) >DroSim_CAF1 9225 112 - 1 CGUGCAA-UGGCGCUCUGGUUGCACGGCGCAGUUGAUGCGAUUACAGCUG-------GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCC (.(((..-.(((((((.((..((.(((((((((((.........))))).-------)).)))).)).)).))).))))....))).)((((..(.((((((.....)))))).).)))) ( -40.40) >DroYak_CAF1 8546 118 - 1 CGUGCAUUUGGUGCU--GUGUGCACGGCGCAAUUGAUGUGAUUACAGCUGCGUGCUGGGUGCUGUGCACUCGAGUUGCCUUAAGCGGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCC (.(((.(..((((((--(.(((((((((((((((.....)))).((((.....)))).))))))))))).).)).))))..).))).)((((..(.((((((.....)))))).).)))) ( -46.50) >consensus CGUGCAA_UGGUGCU_UGGUUGCACGGCGCAGUUGAUGCGAUUACAGCUG_______GCUGCUGGGCUCUCGAGUUGCCUUAAGCAGGGGGCUUUGUUAAAGGAUUACUUUAAGAUGCCC (((((((............))))))).((((.....))))..................(((((((((.........))))..))))).((((..(.((((((.....)))))).).)))) (-32.80 = -32.55 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:43 2006