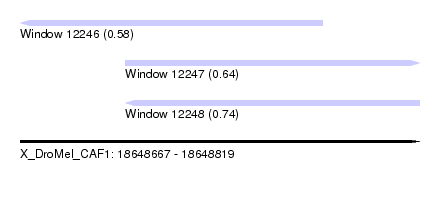
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,648,667 – 18,648,819 |
| Length | 152 |
| Max. P | 0.742969 |

| Location | 18,648,667 – 18,648,782 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -29.86 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18648667 115 - 22224390 UUGGGGUC-----GGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUUCACUGUAGAUCGAAUCAACAAACAUUUUUACAGACUUUCCUGGCCAACG ((((..((-----(((((((((.(((...))).))).))))))))....(((((((((.((..((((......)))).))))).))))))...........(((......))).)))).. ( -41.10) >DroSec_CAF1 7573 119 - 1 UUGGGGUCGUUUUGGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUCCACUGUAGAUCGAAUCAACAAACAUUUUUACAGACUUU-CUGGCCAACG ....((((((((((((((((((.(((...))).))).)))))).)))))(((((((((.((..((((......)))).))))).))))))...................-..)))).... ( -42.20) >DroSim_CAF1 7623 119 - 1 UUGGGGUCGUUUUGGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGCUGAUCGACUUGGGCAGGCUCCACUGUAGAUCGAAUCAACAAACAUUUUUACAGACUUU-CUGGCCAACG .(((((((((((((((((((((.(((...))).))).)))))).)))))(((.(......).))).)))))))(((((((.................))))))).....-.......... ( -40.93) >DroEre_CAF1 7010 114 - 1 UUGGGGUC-----GGCCAUCGUGAGUCAAGCUGACGUAAGGCCAAAAGCGUCGAUCGACUCGGCCAGGCUCCACUGUAGAUCGAGUCAACAAACAUUUUUACACACUUU-CUGGCCAACU ...(((((-----((((..(((.((.....)).)))...))))......(.....))))))(((((((......((((((....((......))...)))))).....)-)))))).... ( -34.30) >DroYak_CAF1 7116 114 - 1 GUGAUGUC-----CGCCUUCGUGAGUGAAGCUGACGUAAGGCAAAAGGCGCCGAUCGACUUGGGCAGGCUCCACUGUAGAUCGAGUCAACAAACAUUUUUACAGACUUU-CUGGCCAACU ........-----.((((((((.((.....)).))).)))))....(((.......(((((((((((......))))...)))))))..............(((.....-)))))).... ( -34.60) >consensus UUGGGGUC_____GGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUCCACUGUAGAUCGAAUCAACAAACAUUUUUACAGACUUU_CUGGCCAACG ....((((.....(((((((((.(((...))).))).))))))......(((((((((.((..((((......)))).))))).))))))......................)))).... (-29.86 = -30.82 + 0.96)

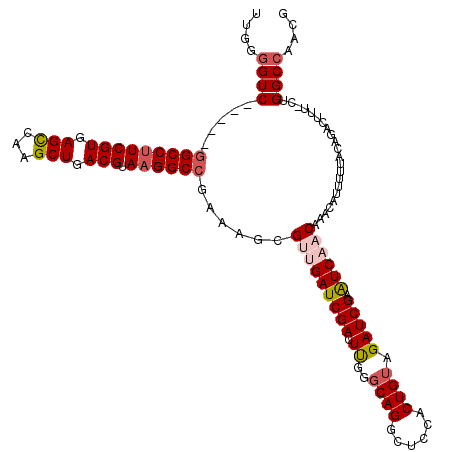
| Location | 18,648,707 – 18,648,819 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -42.14 |
| Consensus MFE | -32.78 |
| Energy contribution | -31.54 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18648707 112 + 22224390 UCUACAGUGAAGCCUGCCCAAGUCGAUCAACGCUUUCGGCCUUACGUCAGCUUGGCUCACGAAGGCC-----GACCCCAAAAGU---AAUGGGAGACAGCUGGUUCAGCUGGGCGAAGCU ..........(((.(((((..(((...........((((((((.(((.(((...))).)))))))))-----)).((((.....---..)))).)))(((((...))))))))))..))) ( -42.80) >DroSec_CAF1 7612 117 + 1 UCUACAGUGGAGCCUGCCCAAGUCGAUCAACGCUUUCGGCCUUACGUCAGCUUGGCUCACGAAGGCCAAAACGACCCCAAAAGU---AAUGGGAGACAGCUGGUUCAGCUGGGCGAAGCU ..........(((.(((((..(((......((.(((.((((((.(((.(((...))).))))))))).)))))..((((.....---..)))).)))(((((...))))))))))..))) ( -41.00) >DroSim_CAF1 7662 117 + 1 UCUACAGUGGAGCCUGCCCAAGUCGAUCAGCGCUUUCGGCCUUACGUCAGCUUGGCUCACGAAGGCCAAAACGACCCCAAAAGU---AAUGGGAGACAGCUGGUUCAGCUGGGCGAAGCU ..........(((.((((((.((.(((((((......((((((.(((.(((...))).)))))))))........((((.....---..)))).....)))))))..))))))))..))) ( -42.80) >DroEre_CAF1 7049 112 + 1 UCUACAGUGGAGCCUGGCCGAGUCGAUCGACGCUUUUGGCCUUACGUCAGCUUGACUCACGAUGGCC-----GACCCCAAAAUG---AAUGGCAGACAGCUGGCUCGGUUGGGCGAAGCA ......((...(((..((((((((...........((((((...(((.((.....)).)))..))))-----))..........---...(((.....)))))))))))..)))...)). ( -41.50) >DroYak_CAF1 7155 115 + 1 UCUACAGUGGAGCCUGCCCAAGUCGAUCGGCGCCUUUUGCCUUACGUCAGCUUCACUCACGAAGGCG-----GACAUCACAAUCUGUAGUGGGUGACAGCUGGUUCAGUUGGGCAAAGCA ...........((.(((((((.(.((.(((((.....))))......(((((((((((((....(((-----((........))))).)))))))).)))))).))).)))))))..)). ( -42.60) >consensus UCUACAGUGGAGCCUGCCCAAGUCGAUCAACGCUUUCGGCCUUACGUCAGCUUGGCUCACGAAGGCC_____GACCCCAAAAGU___AAUGGGAGACAGCUGGUUCAGCUGGGCGAAGCU ......((...(((..((((.((......))......((((((.(((.((.....)).)))))))))......................))))...((((((...)))))))))...)). (-32.78 = -31.54 + -1.24)

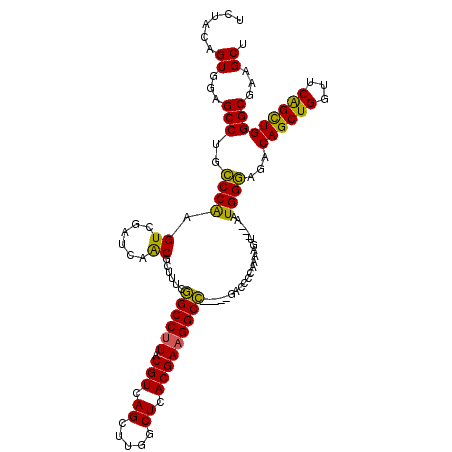

| Location | 18,648,707 – 18,648,819 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -43.34 |
| Consensus MFE | -33.56 |
| Energy contribution | -34.36 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18648707 112 - 22224390 AGCUUCGCCCAGCUGAACCAGCUGUCUCCCAUU---ACUUUUGGGGUC-----GGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUUCACUGUAGA (((((.(((((((((...)))))((((((((..---.....)))))((-----(((((((((.(((...))).))).))))))))...........)))..))))))))).......... ( -47.70) >DroSec_CAF1 7612 117 - 1 AGCUUCGCCCAGCUGAACCAGCUGUCUCCCAUU---ACUUUUGGGGUCGUUUUGGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUCCACUGUAGA (((((.(((((((((...)))))((((((((..---.....))))).(((((((((((((((.(((...))).))).)))))).))))))......)))..))))))))).......... ( -48.70) >DroSim_CAF1 7662 117 - 1 AGCUUCGCCCAGCUGAACCAGCUGUCUCCCAUU---ACUUUUGGGGUCGUUUUGGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGCUGAUCGACUUGGGCAGGCUCCACUGUAGA (((((.(((((((((...)))))(((.((((..---.....))))(.(((((((((((((((.(((...))).))).)))))).))))))).....)))..))))))))).......... ( -48.50) >DroEre_CAF1 7049 112 - 1 UGCUUCGCCCAACCGAGCCAGCUGUCUGCCAUU---CAUUUUGGGGUC-----GGCCAUCGUGAGUCAAGCUGACGUAAGGCCAAAAGCGUCGAUCGACUCGGCCAGGCUCCACUGUAGA ..............(((((.........(((..---.....)))((((-----((((..(((.((.....)).)))...))))....(.(((....))).))))).)))))......... ( -35.10) >DroYak_CAF1 7155 115 - 1 UGCUUUGCCCAACUGAACCAGCUGUCACCCACUACAGAUUGUGAUGUC-----CGCCUUCGUGAGUGAAGCUGACGUAAGGCAAAAGGCGCCGAUCGACUUGGGCAGGCUCCACUGUAGA .((((.((((((......(((((.((((.(((....((..(((.....-----)))..))))).))))))))).((...(((.......)))...))..))))))))))........... ( -36.70) >consensus AGCUUCGCCCAGCUGAACCAGCUGUCUCCCAUU___ACUUUUGGGGUC_____GGCCUUCGUGAGCCAAGCUGACGUAAGGCCGAAAGCGUUGAUCGACUUGGGCAGGCUCCACUGUAGA .((((.((((((.(((....(((...(((((..........))))).......(((((((((.(((...))).))).))))))...))).....)))..))))))))))........... (-33.56 = -34.36 + 0.80)

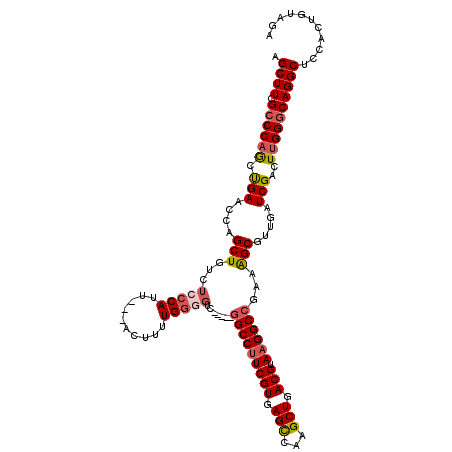
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:39 2006