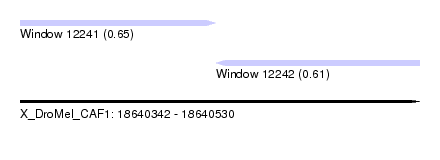
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,640,342 – 18,640,530 |
| Length | 188 |
| Max. P | 0.650696 |

| Location | 18,640,342 – 18,640,434 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18640342 92 + 22224390 ----UUGUGUUUGCCCAAACAUUUUAUGGACGAGUAAAUAGAAAGCUCGCCGAACGUGGGUGGCAUUGUCAUACAUAACCAACCACGGUGAUUUUU------------- ----..((((((....))))))....(((.(((((.........))))))))..(((((.(((...((.....))...))).))))).........------------- ( -23.80) >DroSec_CAF1 15683 94 + 1 --CGUUGUGUUUGCCCAAACAUUUCAUGGACGAGUAAAUAGAAAGCUCGCCGAACGUGGGUGGCAUUGUCAUACAUAACCAACUACGGUGAUUUUC------------- --.(..((((((....))))))..).(((.(((((.........))))))))..(((((.(((...((.....))...))).))))).........------------- ( -23.40) >DroSim_CAF1 16560 94 + 1 --CGUUGUGUUUGCCCAAACAUUUCAUGGACGAGUAAAUAGAAAGCUCGCCGAACGUGGGUGGCAUUGUCAUACAUAACCAACCACGGUGAUUUUC------------- --.(..((((((....))))))..).(((.(((((.........))))))))..(((((.(((...((.....))...))).))))).........------------- ( -25.70) >DroEre_CAF1 16175 107 + 1 --CGUUGUGUUUGCCCAAACAUUUCAUGGAAGAGUAAAUAGAAAGCUCGCGGAACGUGGGUGGCAUUGUCACACAUAGCCGCCCACGUAGAUUUUCCGUAUUUCCCGGA --.(..((((((....))))))..)..((((((((.........))))((((((((((((((((..((.....))..)))))))))))......)))))..)))).... ( -39.40) >DroYak_CAF1 17341 106 + 1 CGCGUUGUGUUUGCCCAAACAUUUCAUGGAAGAGUAAAUAGAAAGCUCUGCGAACGUGGGUGGCAUUGUCAUACAUAACCACCCACGUAGACUUUC-GACUUUCGCA-- .(((..((((((....))))))..)...((.((((.....(((((.(((....((((((((((...((.....))...))))))))))))))))))-.)))))))).-- ( -35.70) >consensus __CGUUGUGUUUGCCCAAACAUUUCAUGGACGAGUAAAUAGAAAGCUCGCCGAACGUGGGUGGCAUUGUCAUACAUAACCAACCACGGUGAUUUUC_____________ ...(..((((((....))))))..).(((.(((((.........))))))))..(((((.(((...((.....))...))).)))))...................... (-19.96 = -20.20 + 0.24)


| Location | 18,640,434 – 18,640,530 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18640434 96 - 22224390 CUCGUU--CGCUCUCCUCCCGGCUGUUCGUGCCCGCAUCCUGG--CGUAAGCAGCAGCUGACAAAGCUCCGGAAAA----GGGGGAAAAGCUCC------GGGAAAUGCG---------- ((((..--.(((.(((.(((.((((((..((((........))--))..))))))((((.....))))........----))))))..)))..)------))).......---------- ( -34.70) >DroSec_CAF1 15777 96 - 1 CUCGCU--CGCUCUCCUCCCGGCUGUUCGUGCCCGCAUCCUGG--CGUGAGCAGCAGCUGACAAAGCUCCGGAAAA----GGGGGAAAAGCUCC------GGGAAAUGCG---------- ((((..--.(((.(((.(((.((((((((((((........))--))))))))))((((.....))))........----))))))..)))..)------))).......---------- ( -37.10) >DroSim_CAF1 16654 96 - 1 CUCGUU--CGCUCUUCUCCCGGCUGUUCGUGUCCGCAUCCUGG--CGUGAGCAGCAGCUGACAAAGCUCCGGAAAA----GGGGGAAAAGCUCC------GGGAAAUGCG---------- ((((..--.(((.(((((((.((((((((((((........))--))))))))))((((.....))))........----))))))).)))..)------))).......---------- ( -38.20) >DroEre_CAF1 16282 104 - 1 CUCGCU--CGCUCUCCUCCCGGCUGUUCGUGCCCACAUCCUGG--CGCGAGCAGCAGCUGACAAAGCUCCGGAAAA--A-GGGGCAAAAGCUCC------GGGAAAUACG---GAAAAGC ......--.(((.((((((((((((((((((((........))--))))))))))((((......(((((......--.-)))))...)))).)------)))).....)---))..))) ( -45.20) >DroYak_CAF1 17447 106 - 1 CACGUUCGCGCUCUCCUCCCGGCUGUUCGUGCCCGCAUCCUGG--CGCGAGCAGCAGCUGACAAAGCUUCGGAAAAACA-GGGGGGAAAGCUCC------GGGAAAUGCG---GAAAG-- .........(((.(((((((.((((((((((((........))--))))))))))((((.....))))...........-))))))).)))(((------(.......))---))...-- ( -48.80) >DroAna_CAF1 29218 113 - 1 CUCCUC--UGCUCUACGC---UCUGUUCGUUUCUC--GCCAGGAACAAAAGGAGCAGCUGACAAAGCGUCGGAAAAGCUUCGCGCGAAAGCCCGCCCACGCUGAAAUCCUGGCGAAAAGC ......--.((.....))---.......((((.((--(((((((......(((((..(((((.....)))))....)))))((((....))..))...........))))))))).)))) ( -37.40) >consensus CUCGUU__CGCUCUCCUCCCGGCUGUUCGUGCCCGCAUCCUGG__CGUGAGCAGCAGCUGACAAAGCUCCGGAAAA____GGGGGAAAAGCUCC______GGGAAAUGCG__________ .........(((.((((((..((((((((((((........))..)))))))))).(((.....))).............))))))..)))............................. (-21.26 = -21.65 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:34 2006