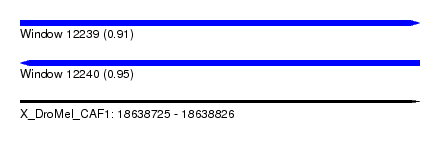
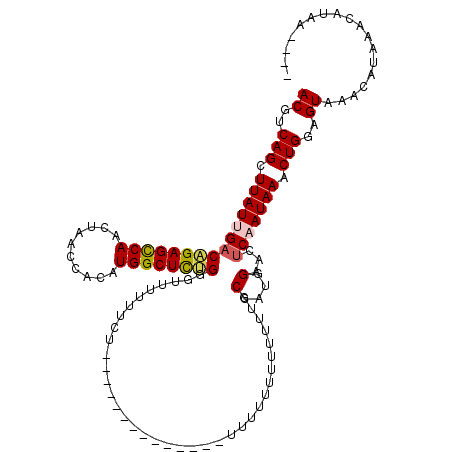
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,638,725 – 18,638,826 |
| Length | 101 |
| Max. P | 0.954415 |

| Location | 18,638,725 – 18,638,826 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -20.66 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18638725 101 + 22224390 ----UUAUGUUUAUGUUUACUCCAGUUUAUUGAGUUCCAUGGCAAAAAAAAAAAAA---------------AAAUACCCCAGAGCCAUGUGGUUAGUUGACUCUGUCCAUAAGCUGACGU ----....(((((((..((....((((.(((((.(..((((((.............---------------............))))))..)))))).)))).))..)))))))...... ( -18.91) >DroSec_CAF1 14054 104 + 1 -----UAUGCUUAUGUUUACUCCAGCUUAUUGAGUUCCAUGGCAAAAAAAAAAAAAGA-----------AGAAAAAACCCAGAGCCAUGUGGUUAGCUGGCUCUGUCUAUAAGCUGACGU -----.................((((((((.((((......))...............-----------..........((((((((.((.....)))))))))))).)))))))).... ( -25.60) >DroSim_CAF1 14938 93 + 1 -----UAUGCUUAUGUUUACUCCAGCUUAUUGAGUUCCAUGGCAAAA----------------------AGAAAAAACCCAGAGCCAUGUGGUUAGCUGGCUCUGUCAAUAAGCUGACGU -----.................(((((((((((((......))....----------------------..........((((((((.((.....))))))))))))))))))))).... ( -29.70) >DroEre_CAF1 14517 103 + 1 CGUUUCCAGUUUCUGUUUACUCCAGUUUAUUGAGUUCCAUGGCGAAAAAAAA-----------------ACAAAAAAUACCGAGCCAAGUAGUUAGUUGGCUCAGUCAAUAAGCUGACGU .((...(((...)))...))..(((((((((((((......)).........-----------------............(((((((.(....).)))))))..))))))))))).... ( -24.60) >DroYak_CAF1 15617 120 + 1 CGUUUCUAGUUUCUGUUUACUCCAGUUUAUUGGGUUCCAUGGCAAAAAAAAAAAAUGAAAAUAAGAGAUACAAAAAAAUCCAAGCCAUGUGGUUAGUUGGCUCUGUCAAUAAGCUGACGU ((((((((((..(((.......)))...))))))...((((((...........((....))..(.(((........))))..)))))).((((.((((((...)))))).)))))))). ( -21.40) >consensus ____UUAUGUUUAUGUUUACUCCAGUUUAUUGAGUUCCAUGGCAAAAAAAAAAAA______________ACAAAAAACCCAGAGCCAUGUGGUUAGUUGGCUCUGUCAAUAAGCUGACGU ......................(((((((((((((......))....................................((((((((..........))))))))))))))))))).... (-20.66 = -21.46 + 0.80)

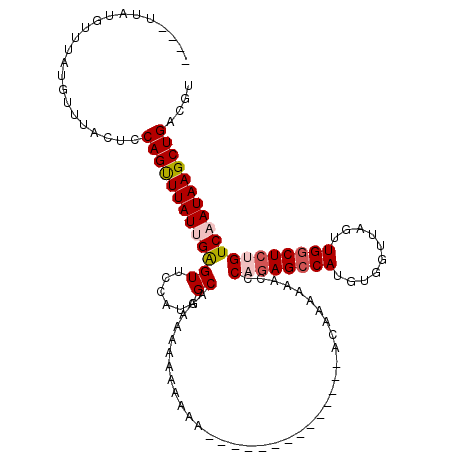
| Location | 18,638,725 – 18,638,826 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18638725 101 - 22224390 ACGUCAGCUUAUGGACAGAGUCAACUAACCACAUGGCUCUGGGGUAUUU---------------UUUUUUUUUUUUUGCCAUGGAACUCAAUAAACUGGAGUAAACAUAAACAUAA---- ((..(((.((((.((((((((((..........)))))))).((((...---------------............)))).......)).)))).)))..))..............---- ( -19.96) >DroSec_CAF1 14054 104 - 1 ACGUCAGCUUAUAGACAGAGCCAGCUAACCACAUGGCUCUGGGUUUUUUCU-----------UCUUUUUUUUUUUUUGCCAUGGAACUCAAUAAGCUGGAGUAAACAUAAGCAUA----- ((..((((((((.((((((((((..........))))))))(((.......-----------...............))).......)).))))))))..)).............----- ( -28.35) >DroSim_CAF1 14938 93 - 1 ACGUCAGCUUAUUGACAGAGCCAGCUAACCACAUGGCUCUGGGUUUUUUCU----------------------UUUUGCCAUGGAACUCAAUAAGCUGGAGUAAACAUAAGCAUA----- ((..(((((((((((((((((((..........))))))))(((.......----------------------....))).......)))))))))))..)).............----- ( -33.20) >DroEre_CAF1 14517 103 - 1 ACGUCAGCUUAUUGACUGAGCCAACUAACUACUUGGCUCGGUAUUUUUUGU-----------------UUUUUUUUCGCCAUGGAACUCAAUAAACUGGAGUAAACAGAAACUGGAAACG ((..(((.((((((((((((((((........)))))))))).......((-----------------(((...........))))).)))))).)))..))...........(....). ( -26.60) >DroYak_CAF1 15617 120 - 1 ACGUCAGCUUAUUGACAGAGCCAACUAACCACAUGGCUUGGAUUUUUUUGUAUCUCUUAUUUUCAUUUUUUUUUUUUGCCAUGGAACCCAAUAAACUGGAGUAAACAGAAACUAGAAACG ((..(((.((((((..((......)).....((((((..(((......((.............)).......)))..)))))).....)))))).)))..)).................. ( -17.04) >consensus ACGUCAGCUUAUUGACAGAGCCAACUAACCACAUGGCUCUGGGUUUUUUCU______________UUUUUUUUUUUUGCCAUGGAACUCAAUAAACUGGAGUAAACAUAAACAUAA____ ((..(((.(((((((((((((((..........)))))))).....................................(....)...))))))).)))..)).................. (-19.06 = -19.30 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:33 2006