
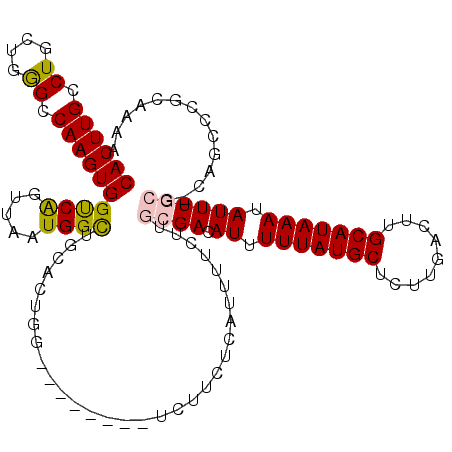
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,634,656 – 18,634,765 |
| Length | 109 |
| Max. P | 0.828663 |

| Location | 18,634,656 – 18,634,765 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18634656 109 + 22224390 CACUUGCCUGCUGGGCCAAGUGGUCAGUUAAUGGCUGCACUGG--------UCUUCUCAUUUUCUUGCGACAUUUUUAUGCUCUUGACUUGCAUAAAUAUUUGCG-CAGCCCGCAAAA ...((((..(((((((((.(..((((.....))))..)..)))--------)).............((((.((.(((((((.........))))))).)))))).-))))..)))).. ( -33.20) >DroPse_CAF1 15105 101 + 1 CACUUGGCUU-AGAGUCAAGUGGCCGGUUAAUGGUGGCAUUGGGGCCUGUCUCUGGGUAUUUUCUUGCGACAUUUUUAUGCUUUCGACUUGCAUAAAUAUUU---------------- ((((((((..-...))))))))((((.....))))(((......)))((((.(.(((.....))).).))))..(((((((.........))))))).....---------------- ( -28.30) >DroSim_CAF1 10957 109 + 1 CACUUGCCUGCUGGGCCAAGUGGUCAGUUAAUGGCUGCACUGG--------UCUUCUCAUUUUCUUGCGACAUUUUUAUGCUCUUGACUUGCAUAAAUAUUUGCG-CAGCCCGCAAAA ...((((..(((((((((.(..((((.....))))..)..)))--------)).............((((.((.(((((((.........))))))).)))))).-))))..)))).. ( -33.20) >DroEre_CAF1 10921 109 + 1 CACUUGCCUGCUGGGCCAAGUGGUCAGUUAAUGGCUGCACUGG--------UCUUCUCAUUUUCUUGCGACAUUUUUAUGCUCUUGACUUGCAUAAAUAUUUGCG-CAGCCCGCAAAA ...((((..(((((((((.(..((((.....))))..)..)))--------)).............((((.((.(((((((.........))))))).)))))).-))))..)))).. ( -33.20) >DroYak_CAF1 11415 110 + 1 CACUUGGCUGCUGGGCCAAGUGGUCAGUUAAUGGCUGCACUGG--------UCUUCUCAUUUUCUUGCGACAUUUUUAUGCUCUUGACUUGCAUAAAUAUUUGCCUUGGCCCGCAAAA ........(((.((((((((.(..((((..........)))).--------.).............((((.((.(((((((.........))))))).)))))))))))))))))... ( -33.40) >DroPer_CAF1 15209 101 + 1 CACUUGGCUU-AGAGUCAAGUGGCCGGUUAAUGGUCGCAUUGGGGCCUGUCUCUGGGUAUUUUCUUGCGACAUUUUUAUGCUUUCGACUUGCAUAAAUAUUU---------------- ((((((((..-...))))))))...........((((((.....((((......)))).......))))))...(((((((.........))))))).....---------------- ( -30.50) >consensus CACUUGCCUGCUGGGCCAAGUGGUCAGUUAAUGGCUGCACUGG________UCUUCUCAUUUUCUUGCGACAUUUUUAUGCUCUUGACUUGCAUAAAUAUUUGCG_CAGCCCGCAAAA ((((((.((....)).))))))((((.....))))...............................((((.((.(((((((.........))))))).)))))).............. (-17.50 = -17.28 + -0.22)


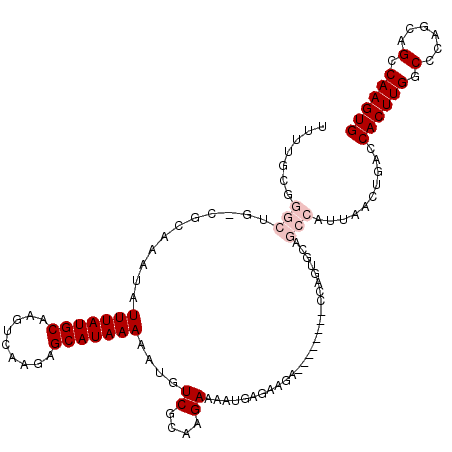
| Location | 18,634,656 – 18,634,765 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18634656 109 - 22224390 UUUUGCGGGCUG-CGCAAAUAUUUAUGCAAGUCAAGAGCAUAAAAAUGUCGCAAGAAAAUGAGAAGA--------CCAGUGCAGCCAUUAACUGACCACUUGGCCCAGCAGGCAAGUG .......(((((-(((.....(((((((.........)))))))....((....))...........--------...))))))))..........((((((.((.....)))))))) ( -33.10) >DroPse_CAF1 15105 101 - 1 ----------------AAAUAUUUAUGCAAGUCGAAAGCAUAAAAAUGUCGCAAGAAAAUACCCAGAGACAGGCCCCAAUGCCACCAUUAACCGGCCACUUGACUCU-AAGCCAAGUG ----------------.....(((((((.........)))))))....((....))...............((((..((((....))))....))))(((((.(...-..).))))). ( -18.80) >DroSim_CAF1 10957 109 - 1 UUUUGCGGGCUG-CGCAAAUAUUUAUGCAAGUCAAGAGCAUAAAAAUGUCGCAAGAAAAUGAGAAGA--------CCAGUGCAGCCAUUAACUGACCACUUGGCCCAGCAGGCAAGUG .......(((((-(((.....(((((((.........)))))))....((....))...........--------...))))))))..........((((((.((.....)))))))) ( -33.10) >DroEre_CAF1 10921 109 - 1 UUUUGCGGGCUG-CGCAAAUAUUUAUGCAAGUCAAGAGCAUAAAAAUGUCGCAAGAAAAUGAGAAGA--------CCAGUGCAGCCAUUAACUGACCACUUGGCCCAGCAGGCAAGUG .......(((((-(((.....(((((((.........)))))))....((....))...........--------...))))))))..........((((((.((.....)))))))) ( -33.10) >DroYak_CAF1 11415 110 - 1 UUUUGCGGGCCAAGGCAAAUAUUUAUGCAAGUCAAGAGCAUAAAAAUGUCGCAAGAAAAUGAGAAGA--------CCAGUGCAGCCAUUAACUGACCACUUGGCCCAGCAGCCAAGUG ((((((((((....)).....(((((((.........)))))))....))))))))...........--------.((((..........))))..((((((((......)))))))) ( -30.90) >DroPer_CAF1 15209 101 - 1 ----------------AAAUAUUUAUGCAAGUCGAAAGCAUAAAAAUGUCGCAAGAAAAUACCCAGAGACAGGCCCCAAUGCGACCAUUAACCGGCCACUUGACUCU-AAGCCAAGUG ----------------.....(((((((.........)))))))....((....))...............((((..((((....))))....))))(((((.(...-..).))))). ( -18.80) >consensus UUUUGCGGGCUG_CGCAAAUAUUUAUGCAAGUCAAGAGCAUAAAAAUGUCGCAAGAAAAUGAGAAGA________CCAGUGCAGCCAUUAACUGACCACUUGGCCCAGCAGCCAAGUG .......(((...........(((((((.........)))))))....((....))...........................)))..........((((((.(......).)))))) (-14.82 = -15.82 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:28 2006