
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,608,026 – 18,608,152 |
| Length | 126 |
| Max. P | 0.615396 |

| Location | 18,608,026 – 18,608,123 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -35.10 |
| Energy contribution | -34.30 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
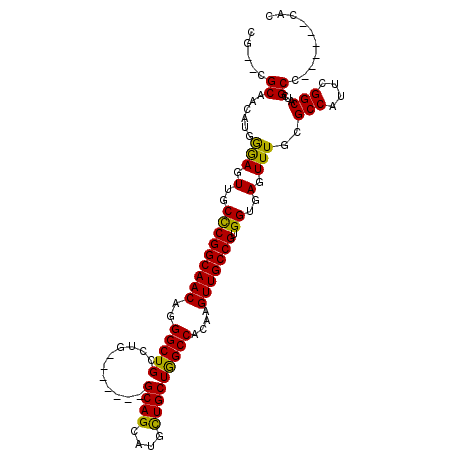
>X_DroMel_CAF1 18608026 97 - 22224390 CG--CGCAACAUGGGAGUUGCCCGGCAACAGGGCUGCCUG-------GCAGCAUGCUGCUAGCCACAAGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCC------CAC ..--.......((((.(.(((((((((((..((((....(-------((((....)))))))))....)))))))(((((......)))))...)))).).))------)). ( -42.90) >DroSec_CAF1 69013 97 - 1 CG--CGCAACAUGGGAGUUGCCCGGCAACAGGGCUGCUUG-------GCAGCAUGCUGCUGGCCACAAGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCC------CAC ..--.......((((.(.(((((((((((..((((....(-------((((....)))))))))....)))))))(((((......)))))...)))).).))------)). ( -42.90) >DroSim_CAF1 67210 97 - 1 CG--CGCAACAUGGGAGUUGCCCGGCAACAGGGCUGCUUG-------GCAGCAUGCUGCUGGCCACAAGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCC------CAC ..--.......((((.(.(((((((((((..((((....(-------((((....)))))))))....)))))))(((((......)))))...)))).).))------)). ( -42.90) >DroEre_CAF1 55644 103 - 1 CG--CGCAACAUGGGAGUUGCUCGGCAACAGGGCUGCCCG-------GCAGGAUGUUGCUGGCCUCAAGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCCCACGCCCAC ..--.......((((.(((((...))))).((((.(.(((-------((((....))))))).)....(.((((((((((......)))))..))))).)))))...)))). ( -41.70) >DroYak_CAF1 60197 106 - 1 CGCGCGCAACAUGAAAGUUGCCCGGCAACAAGGCUGCCCGACAGCAGGCAGCAUGCUGCUGGCCACAUGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCC------CAC .(((.((((((((.....((.((((((.((..((((((........)))))).)).)))))).)))))))))))))((((((....)(((....))).)))))------... ( -46.10) >consensus CG__CGCAACAUGGGAGUUGCCCGGCAACAGGGCUGCCUG_______GCAGCAUGCUGCUGGCCACAAGUUGCCGUGGUGAGUUUGCGCCAUUCGGCAUCGCC______CAC .....((......(((.(..(((((((((..(((((...........((((....)))))))))....))))))).))..).)))..(((....)))...)).......... (-35.10 = -34.30 + -0.80)


| Location | 18,608,055 – 18,608,152 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
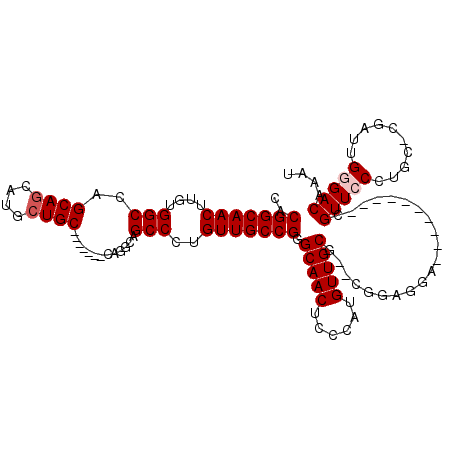
>X_DroMel_CAF1 18608055 97 + 22224390 CACGGCAACUUGUGGCUAGCAGCAUGCUGC-------CAGGCAGCCCUGUUGCCGGGCAACUCCCAUGUUGCG--CGGAGGA----------CGUUCCCUGC-CGAUUGGGACAAAU ..(((((((..(.((((.((.((.....))-------...))))))).)))))))(....)(((((.((((.(--(((.((.----------....))))))-)))))))))..... ( -41.20) >DroSec_CAF1 69042 97 + 1 CACGGCAACUUGUGGCCAGCAGCAUGCUGC-------CAAGCAGCCCUGUUGCCGGGCAACUCCCAUGUUGCG--CGGAGGA----------CGUUCCCUGC-CGAUUGGGACAAAU (((((....)))))(((.((((((.(((((-------...)))))..))))))..)))...(((((.((((.(--(((.((.----------....))))))-)))))))))..... ( -39.70) >DroSim_CAF1 67239 97 + 1 CACGGCAACUUGUGGCCAGCAGCAUGCUGC-------CAAGCAGCCCUGUUGCCGGGCAACUCCCAUGUUGCG--CGGAGGA----------CGUUCCCUGC-CGAUUGGGACAAAU (((((....)))))(((.((((((.(((((-------...)))))..))))))..)))...(((((.((((.(--(((.((.----------....))))))-)))))))))..... ( -39.70) >DroEre_CAF1 55679 98 + 1 CACGGCAACUUGAGGCCAGCAACAUCCUGC-------CGGGCAGCCCUGUUGCCGAGCAACUCCCAUGUUGCG--CGGAGGA----------CGUUCCCUCCGCGAUUGCGACAAAU ..(((((((..(..(((.(((......)))-------..)))..)...))))))).(((((......)))))(--((((((.----------.....)))))))............. ( -42.80) >DroYak_CAF1 60226 117 + 1 CACGGCAACAUGUGGCCAGCAGCAUGCUGCCUGCUGUCGGGCAGCCUUGUUGCCGGGCAACUUUCAUGUUGCGCGCGGAGGAUGUUCUGUUCCGUUCCCUUCCCGAUUGUGACCACU ((((((((((((((.((.((((((.((((((((....))))))))..)))))).)).)).....))))))))(.((((((.(.....).)))))).)..........))))...... ( -48.50) >consensus CACGGCAACUUGUGGCCAGCAGCAUGCUGC_______CAGGCAGCCCUGUUGCCGGGCAACUCCCAUGUUGCG__CGGAGGA__________CGUUCCCUGC_CGAUUGGGACAAAU ..(((((((....(((..((((....)))).............)))..))))))).(((((......))))).....................(((((..........))))).... (-26.86 = -27.46 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:18 2006