
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,604,814 – 18,605,002 |
| Length | 188 |
| Max. P | 0.968156 |

| Location | 18,604,814 – 18,604,927 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -13.24 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18604814 113 + 22224390 AAGAGUUCCUGCCCGAAUGUUGAGCUUCAACUCGUUAAAGUUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA--AAA ..(((((...((.(((...))).))...)))))(((((((((......)))))))))...(((((((..((((((.........))))))...(......)...))))))--).. ( -22.30) >DroSec_CAF1 65852 113 + 1 AAGUUUUCCUGCCCGAAUGUCGAGCUUAAACUCGUUAAAGUUCAUUCAGACUUUAAUGCCUUCGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAA (((((((.(((((....(((((((........((((((((((......))))))))))..)))))))...)))))....)))))))........................--... ( -24.70) >DroSim_CAF1 64074 113 + 1 AAGUUUUCCUGCCCGAAUGUCGAGCUUCAACUCGUUAAAGUUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAA (((((((.(((((....(((((((......)))(((((((((......)))))))))......))))...)))))....)))))))........................--... ( -23.90) >DroYak_CAF1 57188 103 + 1 ------------CUGAAAGUCCAGCUUCAACUUGUUAAAGUUCUUUUAGAACCCAAAGCCUUUGACAACUGCCAACUGAAAAACUUGGCAAACACAAUAAUAAAUGUCGAACAAA ------------(((......))).(((.(((((((((((..((((........)))).))))))))).((((((.(.....).))))))...............)).))).... ( -15.80) >consensus AAGUUUUCCUGCCCGAAUGUCGAGCUUCAACUCGUUAAAGUUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA__AAA .....................(((......)))(((((((..((((........)))).)))))))...((((((.........))))))......................... (-13.24 = -12.80 + -0.44)



| Location | 18,604,854 – 18,604,967 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18604854 113 + 22224390 UUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGAUAAAGGCU ...................((((((....((((((.........))))))....((((.((.(((((...--...))))).))..))))...))))))((((.........)))) ( -19.70) >DroSec_CAF1 65892 113 + 1 UUCAUUCAGACUUUAAUGCCUUCGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCU .................(((((.(.....((((((.........))))))....((((.((.(((((...--...))))).))..))))..................).))))). ( -17.20) >DroSim_CAF1 64114 113 + 1 UUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCU .................(((((((.....((((((.........))))))....((((.((.(((((...--...))))).))..))))..................))))))). ( -21.30) >DroEre_CAF1 52977 99 + 1 GUUUUCCAGACCUU-------------AAUGCCAACUGAAAGACUUGGCAAAC-CAAUCAUGAAUGCUGAACCAAACAUUAAUGCACUCAAAUCAAUGGGGCAAAGGUCA--GCU ........((((((-------------..((((...(((..((.((((....)-))))).(((.(((................))).)))..)))....)))))))))).--... ( -23.29) >DroYak_CAF1 57216 109 + 1 UUCUUUUAGAACCCAAAGCCUUUGACAACUGCCAACUGAAAAACUUGGCAAACACAAUAAUAAAUGUCGAACAAAACAUCAA------CAAAUCAAUGAGGUAAAAACCAAGGCA .................((((((((....((((((.(.....).))))))..............(((.((........)).)------))..)))....(((....)))))))). ( -17.10) >consensus UUCAUUCAGACUUUAAUGCCUUUGACAACUGGCAGCUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA__AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCU .................(((((.......((((((.........))))))............(((((........))))).............................))))). (-10.02 = -10.40 + 0.38)

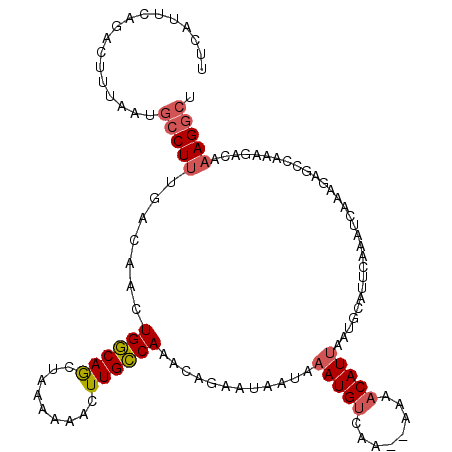
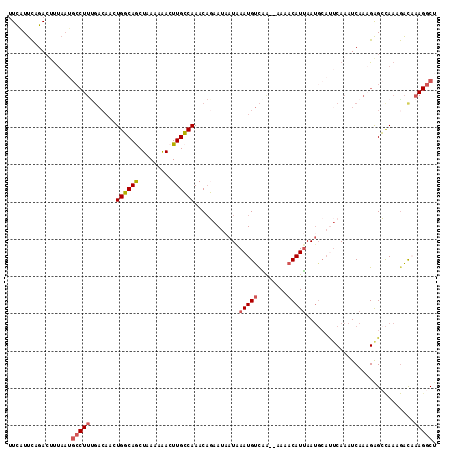
| Location | 18,604,854 – 18,604,967 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18604854 113 - 22224390 AGCCUUUAUCUUUGGCUCUUUGAUUUGAAUGCAUUAAUGUUUU--UUGACAUUUAUUAUUCUGUUUGGCAAGUUUUUUAGCUGCCAGUUGUCAAAGGCAUUAAAGUCUGAAUGAA ((.(((((......((.(((((((..(((((.((.((((((..--..)))))).)))))))...((((((.(((....)))))))))..)))))))))..))))).))....... ( -26.40) >DroSec_CAF1 65892 113 - 1 AGCCUUUGUCUUUGGCUCUUUGAUUUGAAUGCAUUAAUGUUUU--CUGACAUUUAUUAUUCUGUUUGGCAAGUUUUUUAGCUGCCAGUUGUCGAAGGCAUUAAAGUCUGAAUGAA ((.(((((((((((((..........(((((.((.((((((..--..)))))).)))))))...((((((.(((....)))))))))..))))))))))...))).))....... ( -27.70) >DroSim_CAF1 64114 113 - 1 AGCCUUUGUCUUUGGCUCUUUGAUUUGAAUGCAUUAAUGUUUU--CUGACAUUUAUUAUUCUGUUUGGCAAGUUUUUUAGCUGCCAGUUGUCAAAGGCAUUAAAGUCUGAAUGAA ((.(((((((((((((..........(((((.((.((((((..--..)))))).)))))))...((((((.(((....)))))))))..))))))))))...))).))....... ( -28.00) >DroEre_CAF1 52977 99 - 1 AGC--UGACCUUUGCCCCAUUGAUUUGAGUGCAUUAAUGUUUGGUUCAGCAUUCAUGAUUG-GUUUGCCAAGUCUUUCAGUUGGCAUU-------------AAGGUCUGGAAAAC ...--.((((((((((..(((((..(((((((................))))))).(((((-(....)).))))..))))).))))..-------------))))))........ ( -26.89) >DroYak_CAF1 57216 109 - 1 UGCCUUGGUUUUUACCUCAUUGAUUUG------UUGAUGUUUUGUUCGACAUUUAUUAUUGUGUUUGCCAAGUUUUUCAGUUGGCAGUUGUCAAAGGCUUUGGGUUCUAAAAGAA .((((.((((((((((.((.((((.((------((((........))))))...)))).)).).(((((((.(.....).)))))))..)).)))))))..)))).......... ( -23.70) >consensus AGCCUUUGUCUUUGGCUCUUUGAUUUGAAUGCAUUAAUGUUUU__UUGACAUUUAUUAUUCUGUUUGGCAAGUUUUUUAGCUGCCAGUUGUCAAAGGCAUUAAAGUCUGAAUGAA .(((((((...(((((.....((((((........((((((......))))))...............))))))........)))))....)))))))................. (-15.08 = -15.78 + 0.70)


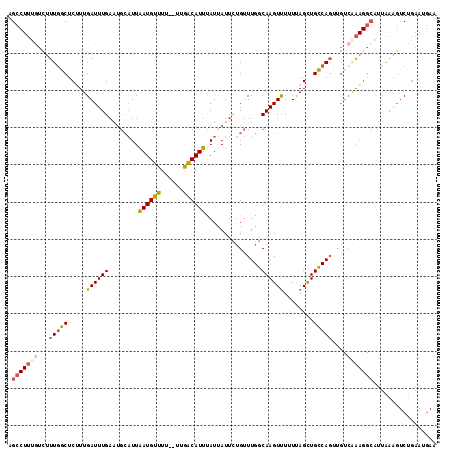
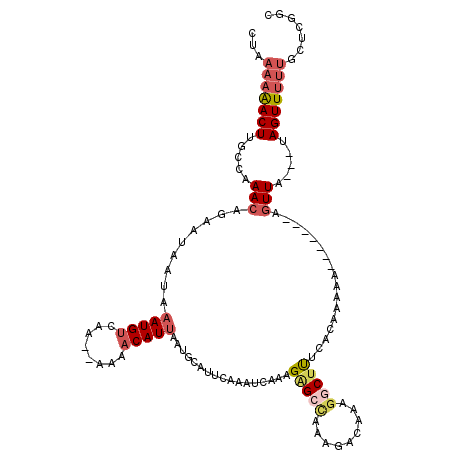
| Location | 18,604,889 – 18,605,002 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.48 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -4.34 |
| Energy contribution | -5.78 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.22 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18604889 113 + 22224390 CUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGAUAAAGGCUUUCACCAAAAAAGUU--AUUUA---UAGUUUUUGCUCGGC ...........(((.....((((.((.(((((...--...))))).))..))))........((((.(((.(((((((((((.......))))))--.))))---)...))).))))))) ( -18.50) >DroSec_CAF1 65927 115 + 1 CUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCUUUCACAAAAAUAUAUAUAGUUA---UAGUUUUUGCUCGGC ...........(((.....((((.((.(((((...--...))))).))..))))........((((.((((((.(..(((.................)))..---).))))))))))))) ( -19.03) >DroSim_CAF1 64149 110 + 1 CUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAG--AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCUUUCACAAAAAA-----AAGUUA---UAGUUUUUGCUCGGC ...........(((.....((((.((.(((((...--...))))).))..))))........((((.((((((.(..(((((........)-----))))..---).))))))))))))) ( -21.70) >DroEre_CAF1 52999 108 + 1 CUGAAAGACUUGGCAAAC-CAAUCAUGAAUGCUGAACCAAACAUUAAUGCACUCAAAUCAAUGGGGCAAAGGUCA--GCUCUCGAAAA---------AGUUAUUGUAGUUAGAGCCUUGA ((((...((((((....)-)))...(((..(((((.((.........(((.((((......)))))))..)))))--))..)))....---------.......))..))))........ ( -19.20) >DroYak_CAF1 57251 103 + 1 CUGAAAAACUUGGCAAACACAAUAAUAAAUGUCGAACAAAACAUCAA------CAAAUCAAUGAGGUAAAAACCAAGGCAUUCGAAAA--------AAGUUA---UAGUUUUUGCCUUGA ........(..((((((.((.(((((.....(((((.......(((.------........)))(((....)))......)))))...--------..))))---).)).))))))..). ( -19.00) >consensus CUAAAAAACUUGCCAAACAGAAUAAUAAAUGUCAA__AAAACAUUAAUGCAUUCAAAUCAAAGAGCCAAAGACAAAGGCUUUCACAAAAA_______AGUUA___UAGUUUUUGCUCGGC ...(((((((.....(((.........(((((........))))).................(((((.........))))).................))).....)))))))....... ( -4.34 = -5.78 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:39:13 2006