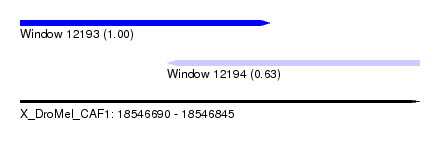
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,546,690 – 18,546,845 |
| Length | 155 |
| Max. P | 0.998876 |

| Location | 18,546,690 – 18,546,787 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.46 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -28.16 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18546690 97 + 22224390 UUAAGGCGGCAUUAAAGGCGAAUUAGCAGAAUUGAUUUGCGCCGGCUUUGCCUUGGGCUGCCUAGAAUUUUUGUGCCCAUAAUCUGGGGAUUAUUCA ...(((((((....((((((((.(.(((((.....)))))....).))))))))..))))))).((((....((.((((.....)))).)).)))). ( -35.40) >DroSec_CAF1 11504 97 + 1 UUAAGGCGGCAUUAAAGGCGAAUUAGCAGAAUUGAUUUGCGCCGGCUUUGCCUUGGGCUGCCUAGAAUUUUUGUGCCCAUAAUCUGGGGAUUAUUCA ...(((((((....((((((((.(.(((((.....)))))....).))))))))..))))))).((((....((.((((.....)))).)).)))). ( -35.40) >DroSim_CAF1 11868 97 + 1 UUAAGGCGGCAUUAAAGGCGAAUUAGCAGAAUUGAUUUGCGCCGGCUUUGCCUUGGGCUGCCUAGAAUUUUUGUGCCCAUAAUCUGGGGAUUAUUCA ...(((((((....((((((((.(.(((((.....)))))....).))))))))..))))))).((((....((.((((.....)))).)).)))). ( -35.40) >DroEre_CAF1 3818 95 + 1 UUAAGACGGUAUUAAAGGCGAAUUAGCCGAAUUGAUUUGCGCCGGCU-UGCCUUGGGCUGCCUAGAAUCUUUGUGCCCAUAAUCUGG-GAUUAUUCA ...((.((((....(((((((.(..((((((....)))).))..).)-))))))..)))).)).((((....((.((((.....)))-))).)))). ( -24.50) >DroYak_CAF1 3977 96 + 1 UUAAGAUGGCAUUAAAGGCGAAUUAGCAGAAUUGAUUUGCGCUGGCU-UGCCUGGGGCUGCCUAGAGUUUUUGUGCCCAUAAUCUGGGGAUUAUUCA .......((((((..((((((.(((((.(((....)))..))))).)-)))))..)).))))..((((....((.((((.....)))).)).)))). ( -28.50) >consensus UUAAGGCGGCAUUAAAGGCGAAUUAGCAGAAUUGAUUUGCGCCGGCUUUGCCUUGGGCUGCCUAGAAUUUUUGUGCCCAUAAUCUGGGGAUUAUUCA ...(((((((....((((((((.(.(((((.....)))))....).))))))))..))))))).((((....((.((((.....)))).)).)))). (-28.16 = -29.08 + 0.92)



| Location | 18,546,747 – 18,546,845 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -10.77 |
| Energy contribution | -11.82 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18546747 98 - 22224390 CUAGAUGGCCCCUUCUC-C------C---A-----UCUACGGCG--UCC----UCCAUCCUCUCCAGCUGCGGAUGCUGUGAAUAAUCCCCAGAUUAUGGGCACAAAAAUUCUAGGCAG (((((((((((......-.------.---.-----..(((((((--(((----..................)))))))))).((((((....)))))))))).)).....))))).... ( -27.97) >DroPse_CAF1 18106 91 - 1 GU-GAUGGCUGAUGCUG-GCUGUC-UGGGA-----UUUGUGCC-----------GCUGCUUCUCUCAAUGCAAA---UAAGCAUAAUCCGAUGAUUGUGCUUGAAGAAA----AAGC-- ..-(((((((......)-))))))-(((((-----...((...-----------...))...))))).......---(((((((((((....)))))))))))......----....-- ( -24.00) >DroSec_CAF1 11561 104 - 1 CUAGAUGGCCCCUUCCC-C------C---A-----UCCACGGCGCAUCCACGGCCCAUCCUCUCCAGCUGCGGACGCUGUGAAUAAUCCCCAGAUUAUGGGCACAAAAAUUCUAGGCAG (((((((((((......-.------.---.-----..(((((((..(((.((((............)))).)))))))))).((((((....)))))))))).)).....))))).... ( -33.70) >DroSim_CAF1 11925 104 - 1 CUAGAUGGCUCCUUCCC-C------C---A-----ACCACGGCGCAUCCACGGCCCAUCCUCUCCAGCUGCGGAUGCUGUGAAUAAUCCCCAGAUUAUGGGCACAAAAAUUCUAGGCAG (((((((((((......-.------.---.-----..((((((..((((.((((............)))).)))))))))).((((((....)))))))))).)).....))))).... ( -29.40) >DroEre_CAF1 3874 83 - 1 CUAGAUGGCUUCUGCCC-A----------------------------------CCCAUCCUCUCCAGCUGCGAAUGCUGUGAAUAAUC-CCAGAUUAUGGGCACAAAGAUUCUAGGCAG (((((...(((.(((((-.----------------------------------.........((((((.......)))).))((((((-...)))))))))))..)))..))))).... ( -24.10) >DroYak_CAF1 4033 110 - 1 CUAGAUGGCUCCUCCACCA-----CC---AGCACCACCAC-CCCCAUUCAUGGCCAAUCCUCUCCAGCUGCGGAUGCUGUGAAUAAUCCCCAGAUUAUGGGCACAAAAACUCUAGGCAG ((((((((........)))-----..---.((.(((....-.....((((((((..((((.(.......).))))))))))))(((((....))))))))))........))))).... ( -25.40) >consensus CUAGAUGGCUCCUUCCC_C______C___A_____UCCACGGCG__UCC____CCCAUCCUCUCCAGCUGCGGAUGCUGUGAAUAAUCCCCAGAUUAUGGGCACAAAAAUUCUAGGCAG (((((((((((...................................................((((((.......)))).))((((((....)))))))))).)).....))))).... (-10.77 = -11.82 + 1.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:51 2006