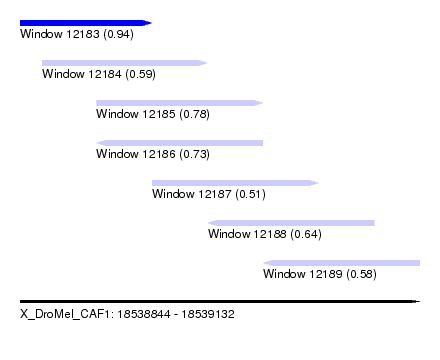
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,538,844 – 18,539,132 |
| Length | 288 |
| Max. P | 0.943555 |

| Location | 18,538,844 – 18,538,939 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -33.79 |
| Energy contribution | -33.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538844 95 + 22224390 ACACCUAAGCUCUAAACUC-GGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUACAUGGAGCAGUCGCCAUGGAGC ........((((((.....-(((((((......(((......((((....)))).....)))((((.(((....))).))))))))))).)))))) ( -31.80) >DroSec_CAF1 3721 96 + 1 ACACCUAAGCUCUAAGCUAAGGCGGCUCAAUGAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCUGUGUCUGCAUGGAGCAGCCGCCAUGGAGC ........((((((.((.((((((((((((((((......))).)))).))))))))).(((((((((((....)))))))).)))))..)))))) ( -38.50) >DroSim_CAF1 3808 96 + 1 ACACCUAAGCUCUAAGCUAAGGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGC ........((((((.((.((((((((((((((...........))))).))))))))).(((((((.(((....))).)))).)))))..)))))) ( -34.60) >consensus ACACCUAAGCUCUAAGCUAAGGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGC ........((((((......(((((((......(((......((((....)))).....)))((((..((....))..))))))))))).)))))) (-33.79 = -33.57 + -0.22)

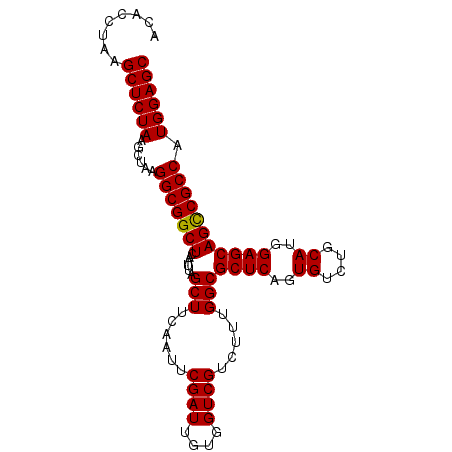

| Location | 18,538,860 – 18,538,979 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -44.72 |
| Energy contribution | -44.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538860 119 + 22224390 CUC-GGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUACAUGGAGCAGUCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCAAACAGCCCAUGUCC ...-(((((((......(((......((((....)))).....)))((((.(((....))).)))))))))))((((.((.((((.....))))((.(....))).....)))))).... ( -42.20) >DroSec_CAF1 3737 120 + 1 CUAAGGCGGCUCAAUGAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCUGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCC (((.(((((((......(((......((((....)))).....)))((((((((....))))))))))))))).)))....((((.....))))(((((..(((......)))..))))) ( -48.60) >DroSim_CAF1 3824 120 + 1 CUAAGGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCC (((.(((((((......(((......((((....)))).....)))((((.(((....))).))))))))))).)))....((((.....))))(((((..(((......)))..))))) ( -45.70) >consensus CUAAGGCGGCUCAAUUAGCUUCAAUUCGAUUGUGGUCGUCUUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCC ....(((((((......(((......((((....)))).....)))((((..((....))..)))))))))))((((.((.((((.....))))((.(....))).....)))))).... (-44.72 = -44.50 + -0.22)


| Location | 18,538,899 – 18,539,019 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -42.69 |
| Energy contribution | -42.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538899 120 + 22224390 UUUGGCGCUCAGUGUCUACAUGGAGCAGUCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCAAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUAGUAGCCAAAUCAA ((((((((((.(((....))).))))....(((((((..(.((((.....))))(((((..(((......)))..)))))..)..))(((.....)))))))).......)))))).... ( -41.60) >DroSec_CAF1 3777 120 + 1 UUUGGCGCUCUGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAA ((((((((((((((....)))))))).((((((((((..(.((((.....))))(((((..(((......)))..)))))..)..))(((.....))))))))...))).)))))).... ( -46.40) >DroSim_CAF1 3864 120 + 1 UUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAA ((((((((((.(((....))).)))).((((((((((..(.((((.....))))(((((..(((......)))..)))))..)..))(((.....))))))))...))).)))))).... ( -43.50) >consensus UUUGGCGCUCAGUGUCUGCAUGGAGCAGCCGCCAUGGAGCAUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAA ((((((((((..((....))..))))(((((((((((..(.((((.....))))(((((..(((......)))..)))))..)..)))......))))..))))......)))))).... (-42.69 = -42.47 + -0.22)

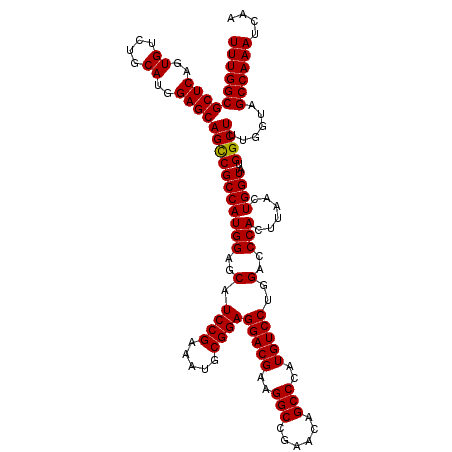
| Location | 18,538,899 – 18,539,019 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -42.33 |
| Energy contribution | -42.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538899 120 - 22224390 UUGAUUUGGCUACUAAAGCCAUACCAGUUAAGUGGGUCCAGGACAUGGGCUGUUUGGCCUUCGUCCUCCGCAUUUCGGAUGCUCCAUGGCGACUGCUCCAUGUAGACACUGAGCGCCAAA ....((((((.......(((((.........((((....(((((..(((((....)))))..))))))))).....((.....)))))))....((((..((....))..)))))))))) ( -42.50) >DroSec_CAF1 3777 120 - 1 UUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCAGGACAUGGGCUGUUCGGCCUUCGUCCUCCGCAUUUCGGAUGCUCCAUGGCGGCUGCUCCAUGCAGACACAGAGCGCCAAA (((((((((((.....)))))....))))))((((....(((((..(((((....)))))..))))))))).....((.(((((..((....((((.....)))).))..)))))))... ( -43.30) >DroSim_CAF1 3864 120 - 1 UUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCAGGACAUGGGCUGUUCGGCCUUCGUCCUCCGCAUUUCGGAUGCUCCAUGGCGGCUGCUCCAUGCAGACACUGAGCGCCAAA (((((((((((.....)))))....))))))((((....(((((..(((((....)))))..))))))))).....((.(((((..((....((((.....)))).))..)))))))... ( -43.20) >consensus UUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCAGGACAUGGGCUGUUCGGCCUUCGUCCUCCGCAUUUCGGAUGCUCCAUGGCGGCUGCUCCAUGCAGACACUGAGCGCCAAA ....((((((.......(((((.........((((....(((((..(((((....)))))..))))))))).....((.....)))))))....((((..((....))..)))))))))) (-42.33 = -42.33 + 0.00)



| Location | 18,538,939 – 18,539,059 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -38.83 |
| Energy contribution | -39.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538939 120 + 22224390 AUCCGAAAUGCGGAGGACGAAGGCCAAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUAGUAGCCAAAUCAAUAACACCCGAUAAGCCGAUUGGUCGAGGGUAAUGGUGGCC .((((.....))))(((((..(((......)))..))))).....(((((....((((.(((((.....))))).)))).....((((.....(((....)))...))))...))))).. ( -38.60) >DroSec_CAF1 3817 120 + 1 AUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAAUAACACCCGAUAAGCCGACUGGGCGAGGGUAAUGGUGGCC .((((.....))))(((((..(((......)))..))))).....(((((....((((.(((((.....))))).)))).....((((.....(((.....)))..))))...))))).. ( -41.40) >DroSim_CAF1 3904 120 + 1 AUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAAUAACACCCGAUAAGCCGACUGGGCGAGGGUAAUGGUGGCC .((((.....))))(((((..(((......)))..))))).....(((((....((((.(((((.....))))).)))).....((((.....(((.....)))..))))...))))).. ( -41.40) >consensus AUCCGAAAUGCGGAGGACGAAGGCCGAACAGCCCAUGUCCUGGACCCACUUAACUGGUAUGGCUUUGGUAGCCAAAUCAAUAACACCCGAUAAGCCGACUGGGCGAGGGUAAUGGUGGCC .((((.....))))(((((..(((......)))..))))).....(((((....((((.(((((.....))))).)))).....((((.....(((.....)))..))))...))))).. (-38.83 = -39.17 + 0.33)



| Location | 18,538,979 – 18,539,099 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -37.51 |
| Energy contribution | -37.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18538979 120 - 22224390 UUUUUGGAAAUCCGCGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGACCAAUCGGCUUAUCGGGUGUUAUUGAUUUGGCUACUAAAGCCAUACCAGUUAAGUGGGUCCA ....((((..(((((.(((((...((((((((....)))))))).....(((((((((....))))......))))).)))))...(((((.....)))))..........))))))))) ( -37.80) >DroSec_CAF1 3857 120 - 1 UUUUUGGAAAUUCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUAUUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCA ....((((...((((.(((((...((((((((....)))))))).....(((((..(((.....))).....))))).)))))...(((((.....)))))..........)))).)))) ( -38.10) >DroSim_CAF1 3944 120 - 1 UUUUUGGAAAUCCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUAUUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCA ....((((..(((((.(((((...((((((((....)))))))).....(((((..(((.....))).....))))).)))))...(((((.....)))))..........))))))))) ( -40.60) >consensus UUUUUGGAAAUCCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUAUUGAUUUGGCUACCAAAGCCAUACCAGUUAAGUGGGUCCA ....((((...((((.(((((...((((((((....)))))))).....(((((..(((.....))).....))))).)))))...(((((.....)))))..........)))).)))) (-37.51 = -37.40 + -0.11)


| Location | 18,539,019 – 18,539,132 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.05 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.03 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18539019 113 - 22224390 AACGAUGGUGGGCAAGUAGAUUCAAUUUCGAAAUUUUUGGAAAUCCGCGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGACCAAUCGGCUUAUCGGGUGUUA (((((((((.((((.(..((((....(((((.....)))))))))..)....)))).((((((((....))))))))))))))((((((((....))))......))))))). ( -36.50) >DroSec_CAF1 3897 113 - 1 AACGAUGGUGGGCAAGCAGAUUCAAUUUCGAAAUUUUUGGAAAUUCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUA (((((((((.((((.((......((((((.((....)).))))))...))..)))).((((((((....))))))))))))))((((..(((.....))).....))))))). ( -38.00) >DroSim_CAF1 3984 113 - 1 AACGAUGGUGGGCAAGUAGAUUCAAUUUCGAAAUUUUUGGAAAUCCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUA (((((((((.((((.((.((((....(((((.....))))))))).))....)))).((((((((....))))))))))))))((((..(((.....))).....))))))). ( -38.10) >consensus AACGAUGGUGGGCAAGUAGAUUCAAUUUCGAAAUUUUUGGAAAUCCACGCAGUGCCAGGUCAUCGCAUGCGAUGGCCACCAUUACCCUCGCCCAGUCGGCUUAUCGGGUGUUA (((((((((.((((.((...(((......))).....(((....))).))..)))).((((((((....))))))))))))))((((..(((.....))).....))))))). (-34.36 = -34.03 + -0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:47 2006