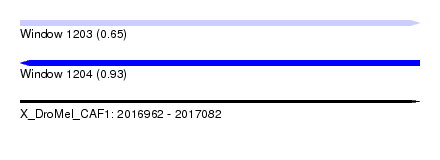
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,016,962 – 2,017,082 |
| Length | 120 |
| Max. P | 0.926186 |

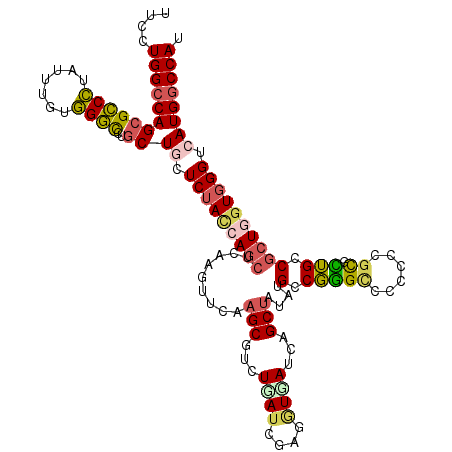
| Location | 2,016,962 – 2,017,082 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -27.01 |
| Energy contribution | -26.99 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2016962 120 + 22224390 GUGGCCAUGGCCGACCAGCGGCAGCACGGGAGGACCCGGCAUGUAGCUGAUGAGGUCAAUCAGCCGCUUGAAUUUUAGAUGGUAGAGCAGCAGGCCCACGAAGAGGGCGCUGGCCAGGAU .(((((((((((...((((.(((((.((((....)))))).))).))))....)))))....((.((((..(((......))).)))).))..((((.......))))..)))))).... ( -48.90) >DroVir_CAF1 7147 120 + 1 AUGGCCAUGACCCAAUAGCGGCCACGCAGGCGGGCUGGCCAAAUCGCGAGUCACCUCGAUGAGAUGCUUGAACUUCAGCUGGUAGAUGAGCAAACCCAAAAAGAGGGCGCUGGCCAAGAA .((((((((.(((......(((((.((......))))))).....((..((((((....((((.(......).))))...))).)))..)).............))))).)))))).... ( -39.90) >DroPse_CAF1 6411 120 + 1 AUGGCCAUGCCCCACCAGCGGCAGAACCGGGGGGCCCGGCAUAUAGCUGAUCACCUCAAUCACACGCUUGAACUUGAGCUGGUAGACCAGCAGGGCCACAAAUAGGCCGCUGGCCAGCAA ...(((..(((((.((...((.....)))))))))..))).....((((((((.((((((((......)))..))))).))))...(((((..((((.......))))))))).)))).. ( -46.80) >DroWil_CAF1 1923 120 + 1 AUGGGCAUGACCCACUAGAGGCAAUGUUGGUGGUCCCGGCAUAUAGCUGAUCACAUCGAUAAUAUGCUUGUAUUUCAACUGAUAGAGUAGUAGGCCAGCAAAUAUGGCGCUGGCCAAAAC .((((.....)))).((.(((((.((((((((....((((.....))))....))))))))...))))).)).....((((......)))).(((((((.........)))))))..... ( -37.50) >DroAna_CAF1 1621 120 + 1 GUGGCCAUGACCCACCAGCGGCAGGGCGGGCGGACCCGGCAUGUAGCUGAUGAUGUUGAUCAGGCGCUUGAACUUGAGCUGGUAGAGCAGCAGGCCCACAAAGAGGGCGCUGGCCAGGAC .((((((((..(.((((((..((((.((((((..((((((.....)))).((((....))))))))))))..)))).)))))).)..))((..((((.......)))))))))))).... ( -47.70) >DroPer_CAF1 6526 120 + 1 AUGGCCAUGCCCCACCAGCGGCAGAACCGGGGGGCCCGGCAUAUAGCUGAUCACCUCAAUCACACGCUUGAACUUGAGCUGGUAGACCAGCAGGGCCACAAAUAGGCCGCUGGCCAGCAA ...(((..(((((.((...((.....)))))))))..))).....((((((((.((((((((......)))..))))).))))...(((((..((((.......))))))))).)))).. ( -46.80) >consensus AUGGCCAUGACCCACCAGCGGCAGAACCGGCGGGCCCGGCAUAUAGCUGAUCACCUCAAUCAGACGCUUGAACUUGAGCUGGUAGAGCAGCAGGCCCACAAAGAGGGCGCUGGCCAGGAA .((((((.........((((........((((....((((.....))))....)))).......)))).........((((......))))..((((.......))))..)))))).... (-27.01 = -26.99 + -0.02)

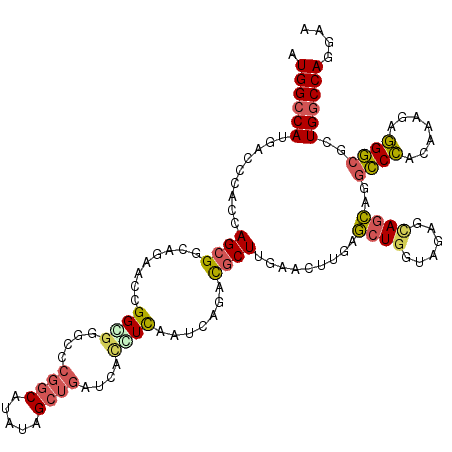
| Location | 2,016,962 – 2,017,082 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -46.22 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2016962 120 - 22224390 AUCCUGGCCAGCGCCCUCUUCGUGGGCCUGCUGCUCUACCAUCUAAAAUUCAAGCGGCUGAUUGACCUCAUCAGCUACAUGCCGGGUCCUCCCGUGCUGCCGCUGGUCGGCCAUGGCCAC .....(((((((((((.......)))).....((...................(..((((((.(....)))))))..)..((((((....)))).)).)).)))))))(((....))).. ( -46.40) >DroVir_CAF1 7147 120 - 1 UUCUUGGCCAGCGCCCUCUUUUUGGGUUUGCUCAUCUACCAGCUGAAGUUCAAGCAUCUCAUCGAGGUGACUCGCGAUUUGGCCAGCCCGCCUGCGUGGCCGCUAUUGGGUCAUGGCCAU ....((((((..((((.......(((((.(((.(((.....(((........))).......((((....)))).)))..))).)))))(((.....))).......))))..)))))). ( -44.80) >DroPse_CAF1 6411 120 - 1 UUGCUGGCCAGCGGCCUAUUUGUGGCCCUGCUGGUCUACCAGCUCAAGUUCAAGCGUGUGAUUGAGGUGAUCAGCUAUAUGCCGGGCCCCCCGGUUCUGCCGCUGGUGGGGCAUGGCCAU ..((((((((((((((.......))))..))))))....((.(((((...((....))...))))).))..)))).....((((.((((((((((......))))).))))).))))... ( -54.80) >DroWil_CAF1 1923 120 - 1 GUUUUGGCCAGCGCCAUAUUUGCUGGCCUACUACUCUAUCAGUUGAAAUACAAGCAUAUUAUCGAUGUGAUCAGCUAUAUGCCGGGACCACCAACAUUGCCUCUAGUGGGUCAUGCCCAU ((...((((((((.......)))))))).))..........((((.....(..((((((....(((...)))....))))))..)......))))..........(((((.....))))) ( -30.10) >DroAna_CAF1 1621 120 - 1 GUCCUGGCCAGCGCCCUCUUUGUGGGCCUGCUGCUCUACCAGCUCAAGUUCAAGCGCCUGAUCAACAUCAUCAGCUACAUGCCGGGUCCGCCCGCCCUGCCGCUGGUGGGUCAUGGCCAC ....((((((((((((.......))))..))((..((((((((.........(((...((((....))))...)))....((.((((......)))).)).))))))))..)))))))). ( -46.40) >DroPer_CAF1 6526 120 - 1 UUGCUGGCCAGCGGCCUAUUUGUGGCCCUGCUGGUCUACCAGCUCAAGUUCAAGCGUGUGAUUGAGGUGAUCAGCUAUAUGCCGGGCCCCCCGGUUCUGCCGCUGGUGGGGCAUGGCCAU ..((((((((((((((.......))))..))))))....((.(((((...((....))...))))).))..)))).....((((.((((((((((......))))).))))).))))... ( -54.80) >consensus UUCCUGGCCAGCGCCCUAUUUGUGGGCCUGCUGCUCUACCAGCUCAAGUUCAAGCGUCUGAUCGAGGUGAUCAGCUAUAUGCCGGGCCCCCCCGCGCUGCCGCUGGUGGGUCAUGGCCAU ....((((((((((((.......))))..))((.(((((((((.........(((...((((....))))...)))....(.(((((......)).))).)))))))))).)))))))). (-29.33 = -29.45 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:43 2006