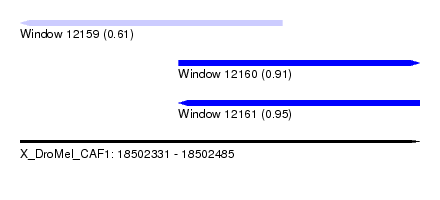
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,502,331 – 18,502,485 |
| Length | 154 |
| Max. P | 0.950226 |

| Location | 18,502,331 – 18,502,432 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -14.61 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18502331 101 - 22224390 ACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUUUUGAAAUGUGUGUUGUAAACACA-------GGCAAGAGCCGCAGAUACAGAUACAUUAACAGACACAG .......(((((........)))))....(((..((.((((((...(((((.......)))))-------..)))))).))..)))...................... ( -20.50) >DroSec_CAF1 10630 97 - 1 ACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUUUUGAAAUGUGUGUUGUAAACACA-------GGCAAGAGUCGCAGAUACAGAUACAUUAACAAAC---- .......(((((........)))))....(((..(((((((((...(((((.......)))))-------..)))))))))..)))..................---- ( -25.50) >DroAna_CAF1 11739 102 - 1 ACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUUUUGAAAUGUGUGUUGUAAACACACACAGGCAGCAAUAGCCGAAGAGUCACACACAC--ACACAC---- .......(((((........)))))..........(((((((....(((((((.(....))))))))(((.......))).)))))))........--......---- ( -25.40) >consensus ACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUUUUGAAAUGUGUGUUGUAAACACA_______GGCAAGAGCCGCAGAUACAGAUACAUUAACAAAC____ .......(((((........)))))....(((..((.((((((.....(((((.....))))).........)))))).))..)))...................... (-14.61 = -15.27 + 0.67)

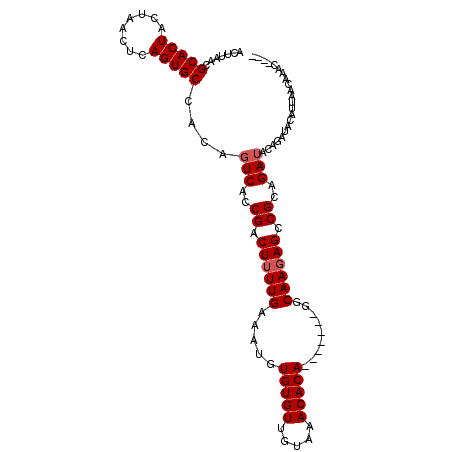

| Location | 18,502,392 – 18,502,485 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -28.76 |
| Energy contribution | -28.72 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18502392 93 + 22224390 AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCAGUCGCGCAGACGCCGACUGAGG--ACGUCGACAGAGACAGCGGA ..(((.(((((((((((((((......))))))(((.....)))...)))))))))(..((((((......))--.))))..)...)))...... ( -35.60) >DroSec_CAF1 10687 93 + 1 AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCAGUCGCGCAGACGCCGACUGAGG--ACGUCGACAGCGGCAGCGGA ...(((.(((((...((((((......))))))....))))).))).(((.(((((...((((((......))--.))))....))))).))).. ( -38.90) >DroEre_CAF1 10761 93 + 1 AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCAGUCGCGCAGACGCCGACCGAGG--ACGUCGACAGAGGCAGCGGG ..(((.(((((((((((((((......))))))(((.....)))...)))))))))...((((((......))--.)))))))............ ( -35.20) >DroYak_CAF1 10787 93 + 1 AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCAGUCGCGCAGACGCCGACAGAGG--ACGUCGACAGAGGCAGCGGG ..(((.(((((((((((((((......))))))(((.....)))...)))))))))...((((((......))--.)))))))............ ( -35.20) >DroMoj_CAF1 18385 95 + 1 AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCUGCCGCGCAGACGCUGACUGCGGCCGCGAUGGCAGCGGCAACAGC ...(((.(((((...((((((......))))))....))))).))).(((((((((((.(.(((.....))).).))).))))))))........ ( -42.20) >consensus AAGUCGGUGACUGUGGCACUGAGUUAGUAGUGCGUUAAGUCAACGAACGCAGUCGCGCAGACGCCGACUGAGG__ACGUCGACAGAGGCAGCGGA ..(((((((.(((((((((((......))))))..........(((......)))))))).))))))).........(((......)))...... (-28.76 = -28.72 + -0.04)



| Location | 18,502,392 – 18,502,485 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18502392 93 - 22224390 UCCGCUGUCUCUGUCGACGU--CCUCAGUCGGCGUCUGCGCGACUGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ...(((((....(((((((.--.(.((((((.((....)))))))).)..))))))).....(((((........))))).)))))......... ( -33.30) >DroSec_CAF1 10687 93 - 1 UCCGCUGCCGCUGUCGACGU--CCUCAGUCGGCGUCUGCGCGACUGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ...((((.....(((((((.--.(.((((((.((....)))))))).)..))))))).....(((((........)))))..))))......... ( -32.10) >DroEre_CAF1 10761 93 - 1 CCCGCUGCCUCUGUCGACGU--CCUCGGUCGGCGUCUGCGCGACUGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ...((((.....(((((((.--.(.((((((.((....)))))))).)..))))))).....(((((........)))))..))))......... ( -31.40) >DroYak_CAF1 10787 93 - 1 CCCGCUGCCUCUGUCGACGU--CCUCUGUCGGCGUCUGCGCGACUGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ..(((.((...((((((((.--....))))))))...))))).......(((.(((((....(((((........)))))...))))).)))... ( -28.80) >DroMoj_CAF1 18385 95 - 1 GCUGUUGCCGCUGCCAUCGCGGCCGCAGUCAGCGUCUGCGCGGCAGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ((....))(((((((..(((((.(((.....))).))))).))))))).(((.(((((....(((((........)))))...))))).)))... ( -40.40) >consensus CCCGCUGCCUCUGUCGACGU__CCUCAGUCGGCGUCUGCGCGACUGCGUUCGUUGACUUAACGCACUACUAACUCAGUGCCACAGUCACCGACUU ..(((.((...(((((((.........)))))))...))))).......(((.(((((....(((((........)))))...))))).)))... (-25.58 = -25.54 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:23 2006