
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,500,450 – 18,500,553 |
| Length | 103 |
| Max. P | 0.811134 |

| Location | 18,500,450 – 18,500,553 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.95 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
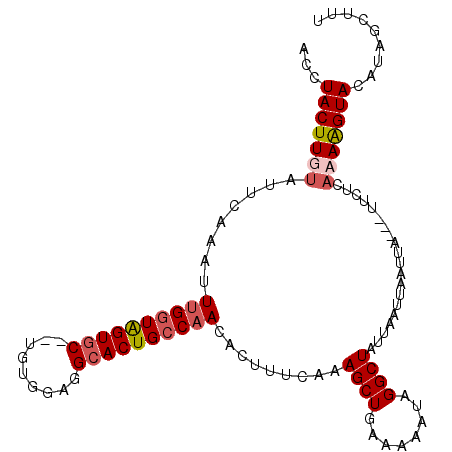
>X_DroMel_CAF1 18500450 103 + 22224390 ACCUACUUGUUUUCAAAUUUGGUAGUGC--UGUGGCGGCACUGCCAACACUUUCCGAGCUGAUAAAUAGGCUAUUAAUAAGUUA---UUCUUUAAAGUACAUAGCUUU ....(((((((.......((((((((((--((...)))))))))))).........((((........))))...)))))))..---......(((((.....))))) ( -25.50) >DroSec_CAF1 8653 103 + 1 ACCUACUUGUAUUCAAAUUUGGUAGUAC--UGUGGCGGCACUGCCAACACUUUCCAAGCUGAAAAGUAGGCUGUUAAUUAGUUA---UUCUUAAAAGUACAUAGCUUU .(((((((...((((..(((((.(((..--..(((((....)))))..)))..))))).))))))))))).........(((((---(.((....))...)))))).. ( -23.30) >DroEre_CAF1 8809 101 + 1 ACCUACUUGUAUUCAAAUUUGGUAGUGCCGUGUGGAGGCACUGCCAACGCUUUCAAAGCUGAAAAAUAGGCUAUUAAUUAAUUAUUAUUCGCAUAAGUAUA------- ...(((((((.((((...(((((((((((.......))))))))))).(((.....)))))))......((...((((.....))))...)))))))))..------- ( -29.50) >DroYak_CAF1 8793 103 + 1 ACCUACUUGUAUUCAAAUUAGUUGGUGC--UGUGGAGGCACUGCCCACACUUUCAAAGCUGAAAAAUAGGCUAUUAAUUAAUUA---UUCUCAUAGGUAUAUGACUUU (((((..((......(((((((((((((--((((..(((...)))..)))(((((....)))))....))).))))))))))).---....))))))).......... ( -22.80) >consensus ACCUACUUGUAUUCAAAUUUGGUAGUGC__UGUGGAGGCACUGCCAACACUUUCAAAGCUGAAAAAUAGGCUAUUAAUUAAUUA___UUCUCAAAAGUACAUAGCUUU ...(((((((........((((((((((.........)))))))))).........((((........))))....................)))))))......... (-15.82 = -16.95 + 1.13)


| Location | 18,500,450 – 18,500,553 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18500450 103 - 22224390 AAAGCUAUGUACUUUAAAGAA---UAACUUAUUAAUAGCCUAUUUAUCAGCUCGGAAAGUGUUGGCAGUGCCGCCACA--GCACUACCAAAUUUGAAAACAAGUAGGU .....................---.............((((((((.((((...((..(((((((((......))))..--))))).))....))))....)))))))) ( -19.00) >DroSec_CAF1 8653 103 - 1 AAAGCUAUGUACUUUUAAGAA---UAACUAAUUAACAGCCUACUUUUCAGCUUGGAAAGUGUUGGCAGUGCCGCCACA--GUACUACCAAAUUUGAAUACAAGUAGGU .....................---.............(((((((((((((.((((..((((.((((......))))..--.)))).))))..)))))...)))))))) ( -21.50) >DroEre_CAF1 8809 101 - 1 -------UAUACUUAUGCGAAUAAUAAUUAAUUAAUAGCCUAUUUUUCAGCUUUGAAAGCGUUGGCAGUGCCUCCACACGGCACUACCAAAUUUGAAUACAAGUAGGU -------..............................(((((((((((((((.....))).((((.((((((.......)))))).))))...))))...)))))))) ( -25.40) >DroYak_CAF1 8793 103 - 1 AAAGUCAUAUACCUAUGAGAA---UAAUUAAUUAAUAGCCUAUUUUUCAGCUUUGAAAGUGUGGGCAGUGCCUCCACA--GCACCAACUAAUUUGAAUACAAGUAGGU ..........((((((.....---.....(((((...(((((((((((......))))).)))))).((((.......--))))....))))).........)))))) ( -21.51) >consensus AAAGCUAUAUACUUAUAAGAA___UAACUAAUUAAUAGCCUAUUUUUCAGCUUGGAAAGUGUUGGCAGUGCCGCCACA__GCACUACCAAAUUUGAAUACAAGUAGGU .....................................(((((((((((((((.....))).((((.(((((.........))))).))))...))))...)))))))) (-18.41 = -18.23 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:21 2006