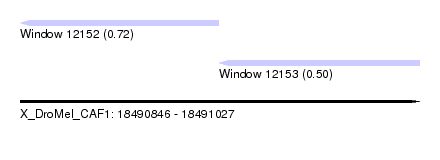
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,490,846 – 18,491,027 |
| Length | 181 |
| Max. P | 0.721188 |

| Location | 18,490,846 – 18,490,936 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -17.73 |
| Energy contribution | -17.56 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18490846 90 - 22224390 UCCCAAUUCC------AAUCCGCAUC--CCAGGCUGCGGCUUCUUCUUCCUAUACGUGCAGCAUGUGCGCGUGAUCUUCGAGACGCGCACCAACAUCC ..........------...(((((.(--....).))))).........................((((((((..((...)).))))))))........ ( -21.20) >DroVir_CAF1 2611 83 - 1 ---------U------AAUACGCGGCCAUUAGGCUCGGGUUUCUUUUUUCUGUAUAUGCACCAUGUGCGCGUCAUCUUUGAGACGCGCACCAACAUUC ---------.------...((.(((((....))).)).))........................(((((((((........)))))))))........ ( -25.40) >DroSec_CAF1 2110 90 - 1 UCCCAAUUCA------AAUCCGCAUC--CCAGGCUGCGGCUUCUUCUUCCUGUACGUGCAGCAUGUGCGCGUGAUCUUCGAGACGCGCACCAACAUCC ..........------...(((((.(--....).)))))..........((((....))))...((((((((..((...)).))))))))........ ( -23.50) >DroEre_CAF1 2089 90 - 1 UCCAAAUUUC------AAUCCGCAUC--CCAGGCUCCGGCUUCUUCUUCCUGUACGUGCAGCAUGUGCGCGUGAUCUUCGAGACGCGCACCAACAUCC ..........------.....((((.--.((((...............))))...)))).....((((((((..((...)).))))))))........ ( -20.26) >DroYak_CAF1 2099 96 - 1 UCCAAAUCUCCAUCUCCAUCCGCAUC--CCAGGCUCCGGCUUCUUCUUCCUGUACGUGCAGCAUGUGCGCGUGAUCUUUGAGACGCGCACCAACAUCC .....................((((.--.((((...............))))...)))).....((((((((..((...)).))))))))........ ( -20.26) >DroMoj_CAF1 2317 83 - 1 ---------U------AAUACGCAGCCAUUAGGCUCGGGUUUCUUUUUUCUGUAUGUGCACCAUGUGCGCGUUAUCUUCGAGGCGCGCACCAACAUAC ---------.------....((.((((....))))))..............((((((.......(((((((((........)))))))))..)))))) ( -25.70) >consensus UCC_AAUU_C______AAUCCGCAUC__CCAGGCUCCGGCUUCUUCUUCCUGUACGUGCAGCAUGUGCGCGUGAUCUUCGAGACGCGCACCAACAUCC .....................((((....((((...............))))...)))).....((((((((..........))))))))........ (-17.73 = -17.56 + -0.17)


| Location | 18,490,936 – 18,491,027 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18490936 91 - 22224390 UCCGCCAACCGGCAGACACUUCUGCCACGCCUCUUCCGCUUCCAGG-CGGCAGUCUAAUUGAGUUGGGGAUUCCCAC------UGCCAUUGACCACAC ...(((....((((((....))))))..((.......)).....))-)(((((((.....))..((((....)))))------))))........... ( -29.00) >DroSec_CAF1 2200 91 - 1 UCCGCCAACCGGCAUACACUUCUGCCACGCCUCUUCCGCUUCCAGG-CGGCAGUCCAAUUAAGUUGGGGAUUCCCAU------UGCCAUUGACCACAC (((.(((((.((((........))))......((.(((((....))-))).)).........)))))))).......------............... ( -23.60) >DroSim_CAF1 921 91 - 1 UCCGCCAACCGGCAGACACUUCUGCCACGCCUCUUCCGCUUCCAGG-CGGCAGUCCAAUUGAGUUGGGGAUUCCCAU------UGCCAUUGACCACAC ...(((....((((((....))))))..((((...........)))-)))).(..((((.(...((((....)))).------..).))))..).... ( -28.60) >DroEre_CAF1 2179 91 - 1 UCCAUGUACCGGCAGACUGUUCUGCCACGCCUCUUCCGCUUCCAGG-GGACAGUCCAAUUCAAUCGGGGAUUCCCAC------UGCCAUUCACCAAAC ..........(((((((((((((.....((.......))......)-))))))))..........(((....)))..------))))........... ( -22.30) >DroYak_CAF1 2195 98 - 1 UCCAUAAACCGGCAGACUGCUCUGCCACGCCUCUUCCGCUUCCAGGAGGGCAGUCCAAUUCAAUCGGGGAUUCCCAUUGUCAUUGCCAUUGACCAAAC ..........(((((((((((((.((..((.......)).....))))))))))).....((((.(((....)))))))....))))........... ( -27.50) >consensus UCCGCCAACCGGCAGACACUUCUGCCACGCCUCUUCCGCUUCCAGG_CGGCAGUCCAAUUCAGUUGGGGAUUCCCAU______UGCCAUUGACCACAC ..........((((.......(((((...(((...........)))..)))))............(((....)))........))))........... (-17.84 = -18.04 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:16 2006