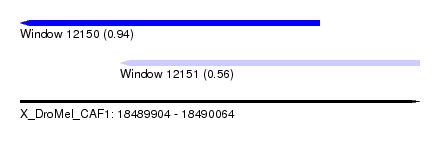
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,489,904 – 18,490,064 |
| Length | 160 |
| Max. P | 0.942589 |

| Location | 18,489,904 – 18,490,024 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.69 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.65 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
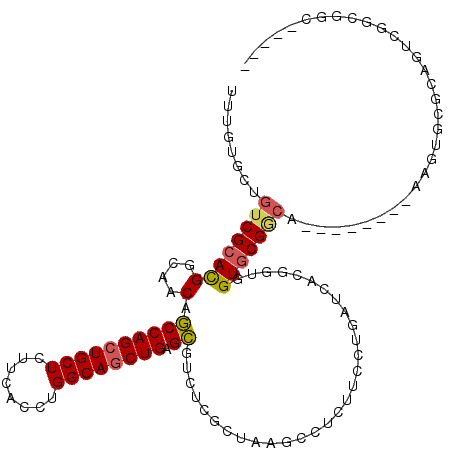
>X_DroMel_CAF1 18489904 120 - 22224390 UUUGUAAUGUCGCACGGCAACAGCCAGCUGCUUCUCACCUGGCAGCUGAGCAUCUCGCUGUUCCUAUUUUUGAUUACAAUGGUGCGACAGUUGCGUCACGUGCGCAGCCGGCGACUGGGA ........(((((.((((....((((((((((........)))))))).)).....((((((.(.((..(((....)))..))).))))))(((((.....))))))))))))))..... ( -46.20) >DroPse_CAF1 1185 106 - 1 UUCGUGCUGUCGCAUGGGAACAGCCAGCUGCUGUUUACCUGGCAGCUGAGCGUGUCGAUAAGCCUCUUCCUUAUCACGGUGGUGCGGC---------AAGUGCGGAGUCGACGGC----- ((((..(((((((((.(((...((((((((((........)))))))).))...))((((((.......))))))....).)))))))---------.))..)))).........----- ( -43.90) >DroGri_CAF1 1119 106 - 1 UUUGUGCUCUCGCAUGGCAACAGCCAGCUGCUUUUCACCUGGCAGCUGAGCGUUGCACUUAGCUUGUUCCUGAUUACGGUUGUUCGGCA---------AUUACGCAAUCGGCGCU----- ...((((....((..((((((.((((((((((........)))))))).))))))).)...)).............((((((..((...---------....)))))))))))).----- ( -36.70) >DroEre_CAF1 1138 120 - 1 UUUGUGAUGUCGCACGGCAACAGCCAGCUGCUCUUUACCUGGCAGCUGAGCAUCUCGCUGUUCCUAUUUCUGAUUACAGUGGUGCGACAAUUGCGUUAUGUGCGCAACCGGCGGCUGGGA .......(((((((((....).((((((((((........)))))))).))....((((((..............)))))))))))))).((((((.....))))))((((...)))).. ( -47.24) >DroAna_CAF1 1126 120 - 1 UUUAUCCUGUCGCACGGCAACAACCAGCUGCUCUUCACCUGGCAACUGAGUGUCUCCUUUUCCCUCUUCCUCAUCACGGUGGUGCGUCAUGUGCGCCAAGUGCGCAGUCGUCGGCAGGGC ....((((((((.(((((.(((..(((.((((........)))).)))..))).....................(((..(((((((.....))))))).)))....))))))))))))). ( -41.40) >DroPer_CAF1 1179 106 - 1 UUCGUGCUGUCGCAUGGGAACAGCCAGCUGCUGUUUACCUGGCAGCUGAGCGUGUCGAUAAGCCUCUUCCUUAUCACGGUGGUGCGGC---------AAGUGCGGAGUCGACGGC----- ((((..(((((((((.(((...((((((((((........)))))))).))...))((((((.......))))))....).)))))))---------.))..)))).........----- ( -43.90) >consensus UUUGUGCUGUCGCACGGCAACAGCCAGCUGCUCUUCACCUGGCAGCUGAGCGUCUCGCUAAGCCUCUUCCUGAUCACGGUGGUGCGGCA________AAGUGCGCAGUCGGCGGC_____ ........((((((((....).((((((((((........)))))))).))..............................)))))))................................ (-25.68 = -25.65 + -0.03)

| Location | 18,489,944 – 18,490,064 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
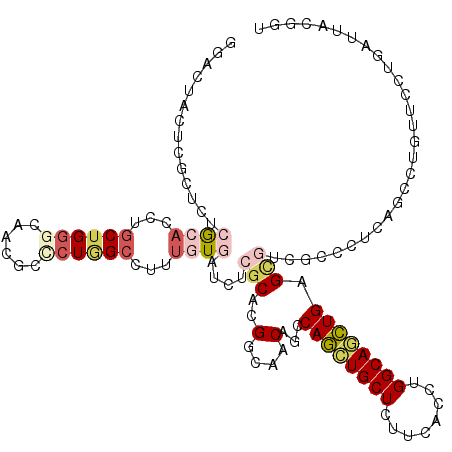
>X_DroMel_CAF1 18489944 120 - 22224390 GGACUACUCGCUGUCGCACCUGCUGGGCCACGCUCUCGCCUUUGUAAUGUCGCACGGCAACAGCCAGCUGCUUCUCACCUGGCAGCUGAGCAUCUCGCUGUUCCUAUUUUUGAUUACAAU (((......(((((((((..(((.((((.........))))..))).)).)).))))).(((((.(((((((........)))))))(((...)))))))))))................ ( -35.50) >DroVir_CAF1 1118 120 - 1 GGACUACUCGCUCUCACAUUUGCUGGGCAAUUGCCUGGCCUUUGUGAUCUCGCACGGCAACACCCAACUGCUCUUCACCUGGCAGUUGAGCGUUGCCCUCAGCCUGUUCCUAAUAACGAU (((..((..(((.(((((...((..(((....)))..))...)))))........((((((.(.((((((((........)))))))).).))))))...)))..))))).......... ( -45.40) >DroPse_CAF1 1211 120 - 1 GGACUACUCGCUGUCGCACCUGCUGGGCAACGCGAUAGCCUUCGUGCUGUCGCAUGGGAACAGCCAGCUGCUGUUUACCUGGCAGCUGAGCGUGUCGAUAAGCCUCUUCCUUAUCACGGU .(((..(((((((((((.((....))))...((((((((......))))))))..(((((((((.....))))))).)).)))))).)))...)))((((((.......))))))..... ( -48.20) >DroGri_CAF1 1145 120 - 1 GGAUUACUCACUGUCACACCUGCUGGGCAAUUGCCUGGCCUUUGUGCUCUCGCAUGGCAACAGCCAGCUGCUUUUCACCUGGCAGCUGAGCGUUGCACUUAGCUUGUUCCUGAUUACGGU (((.........(.((((...((..(((....)))..))...)))))....((..((((((.((((((((((........)))))))).))))))).)...))....))).......... ( -43.00) >DroMoj_CAF1 1187 120 - 1 GGACUAUUCGCUCUCGCAUCUGCUGGGCAACGCCCUGGCCUUUCUGAUCUCACACGGCAACACGCAGUUGCUCUUCACCUGGCAACUGAGCCUGGCCCUCAGCAUGUUCAUGAUUACGCU ((((.....((....))...(((((((....(((..(((................(....)...((((((((........)))))))).))).))).))))))).))))........... ( -35.10) >DroAna_CAF1 1166 120 - 1 UGACUACUCCCUCUCGCACAUACUGGCCCAUGCCCUGGCCUUUAUCCUGUCGCACGGCAACAACCAGCUGCUCUUCACCUGGCAACUGAGUGUCUCCUUUUCCCUCUUCCUCAUCACGGU .........((....(((((((..((((........))))..))...))).))..((..(((..(((.((((........)))).)))..)))..))....................)). ( -22.60) >consensus GGACUACUCGCUCUCGCACCUGCUGGGCAACGCCCUGGCCUUUGUGAUCUCGCACGGCAACAGCCAGCUGCUCUUCACCUGGCAGCUGAGCGUCGCCCUCAGCCUGUUCCUGAUUACGGU ..............((((...((((((......))))))...))))....(((..(....)...((((((((........)))))))).)))............................ (-18.47 = -19.78 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:15 2006