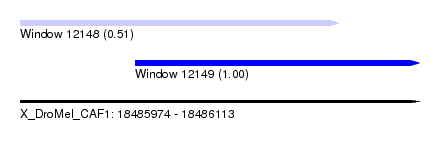
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,485,974 – 18,486,113 |
| Length | 139 |
| Max. P | 0.999293 |

| Location | 18,485,974 – 18,486,085 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -46.93 |
| Consensus MFE | -31.74 |
| Energy contribution | -34.18 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
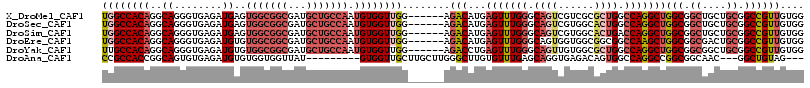
>X_DroMel_CAF1 18485974 111 + 22224390 UGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAUGUGGUUGG------AGACAUGAGUUUGGGCAGUCGUCGCGCUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGG .((((.((..((........))..))((((((..((((((........((.------...))..(((((((.((((......)))).)))))))))))))))))))))))....... ( -48.80) >DroSec_CAF1 3822 111 + 1 UGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAUGUGGUUGG------AGACAUGAGUUUGGGCAGUCGUGGCACUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGG .((((.((..((........))..))((((((..((((((........((.------...))..(((((((.((((......)))).)))))))))))))))))))))))....... ( -48.50) >DroSim_CAF1 3801 111 + 1 UGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAUGUGGUUGG------AGACAUGAGUUUGGGCAGUCGUGGCACUGACCAGGCUGGCGGCUGCUGCGGCCGUUGUGG .((((.((..((........))..))((((((..(((((((.(.((((..(------.(.(((((.(......).))))).).)..)))).).)))))))))))))))))....... ( -47.80) >DroEre_CAF1 4305 111 + 1 UGGCCACAGGCAGGGUGAGAUGUGUGGCGGCGAUGCUGCCAAUGUGGUUGG------AGACAUGAGUUUGGGCAGUGGUGGCGGCGGCCAAGCUGGCGGCGACUGCGGCCGUUGUGG .((((.(((((.............(((((((...))))))).......((.------...))...))))).(((((.((.(((((......))).)).)).)))))))))....... ( -45.90) >DroYak_CAF1 3745 111 + 1 UUGCCACAGGCAGGGUGAGAUGUGUGGCGGCGAUGCUGCCAAUGUGGUUGG------AGACCUGAGUUUGGGCAGUUGUGGCGCUGGCCAGGCUGGCGGCGGCUGCGGCCGUUGUGG ...(((((((((.(......).)))(((.(((.(((((((.....((((..------.))))..(((((((.((((......)))).))))))))))))))..))).))).)))))) ( -49.10) >DroAna_CAF1 3909 102 + 1 CCGCCACCGGCAGUGUGAGAUGUGUGGUGGUUAU---------GUGGUUGCUUGCUUGGGCUUGUGUUUGAGCAGGUGAGACAGUGGCCAGGCCGGCGGCAAC---GGCUGUAG--- ..(((.(((((.(..(.....)..)..(((((((---------....(..((((((..(((....)))..))))))..)....))))))).))))).)))...---........--- ( -41.50) >consensus UGGCCACAGGCAGGGUGAGAUGAGUGGCGGCGAUGCUGCCAAUGUGGUUGG______AGACAUGAGUUUGGGCAGUCGUGGCACUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGG ((((((((..((........))..(((((((...))))))).))))))))........(((...(((((((.((((......)))).)))))))(((.((....)).)))))).... (-31.74 = -34.18 + 2.45)


| Location | 18,486,014 – 18,486,113 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -44.40 |
| Consensus MFE | -42.76 |
| Energy contribution | -42.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18486014 99 + 22224390 AAUGUGGUUGGAGACAUGAGUUUGGGCAGUCGUCGCGCUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGGCGGACGAAGAUGAGGCUGCCGCGGCAGU .....(((((.((((....)))).(((((((((((.(((.....)))))))))))))).)))))((((((((((.(........).))))))))))... ( -44.70) >DroSec_CAF1 3862 99 + 1 AAUGUGGUUGGAGACAUGAGUUUGGGCAGUCGUGGCACUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGGCGGACGAAGAUGAGGCCGCCGCGGCAGU ........((....))..(((((((.((((......)))).)))))))(((.((....)).)))((((((((((.(........).))))))))))... ( -45.60) >DroSim_CAF1 3841 99 + 1 AAUGUGGUUGGAGACAUGAGUUUGGGCAGUCGUGGCACUGACCAGGCUGGCGGCUGCUGCGGCCGUUGUGGCGGACGAAGAUGAGGCCGCCGCGGCAGU .....(((((.((((....)))).((((((((((((.((....))))).))))))))).)))))((((((((((.(........).))))))))))... ( -43.60) >DroEre_CAF1 4345 99 + 1 AAUGUGGUUGGAGACAUGAGUUUGGGCAGUGGUGGCGGCGGCCAAGCUGGCGGCGACUGCGGCCGUUGUGGCGGACGAAGAGGAGGCCGCCGCAGCAGU .....(((((....)..........(((((.((.(((((......))).)).)).)))))))))((((((((((.(........).))))))))))... ( -44.40) >DroYak_CAF1 3785 99 + 1 AAUGUGGUUGGAGACCUGAGUUUGGGCAGUUGUGGCGCUGGCCAGGCUGGCGGCGGCUGCGGCCGUUGUGGCGGACGAGGAGGAGGCCGUCGCGGCAGU .....(((((.((.(((.(((((((.((((......)))).))))))).).))...)).)))))((((((((((.(........).))))))))))... ( -43.70) >consensus AAUGUGGUUGGAGACAUGAGUUUGGGCAGUCGUGGCGCUGGCCAGGCUGGCGGCUGCUGCGGCCGUUGUGGCGGACGAAGAUGAGGCCGCCGCGGCAGU ........((....))..(((((((.((((......)))).)))))))(((.((....)).)))((((((((((.(........).))))))))))... (-42.76 = -42.48 + -0.28)

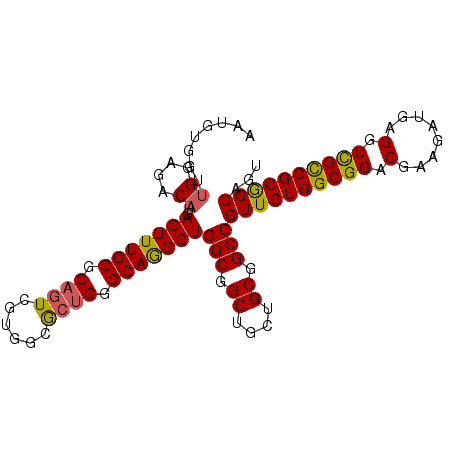
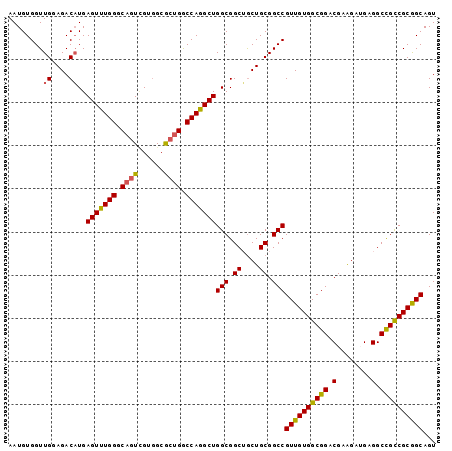
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:13 2006