
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,475,024 – 18,475,141 |
| Length | 117 |
| Max. P | 0.974815 |

| Location | 18,475,024 – 18,475,125 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -24.07 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18475024 101 + 22224390 UUUUU----U--UCGCUUGCACCUCGGCGCCGUCUCCGAUUCUAGCUCAUCGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUC .....----.--.((((........))))..(.(((((((((((((((...(((((((.......)))))))..(((....)))......))))))))))))))).) ( -40.20) >DroSec_CAF1 3708 101 + 1 UUUUU----U--UCGCGUAUACCUCGGCGCCGUCUUCGAUUCCAGCUCACUGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUC .....----.--..((((........)))).(.(((((((((((((((...(((((((.......)))))))..(((....)))......))))))))))))))).) ( -40.80) >DroSim_CAF1 2846 101 + 1 UUUUU----U--UCGCGUAUACCUCGGCGACGUCUUCGAUUCCAGCUCACUGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUC .....----.--((((..........)))).(.(((((((((((((((...(((((((.......)))))))..(((....)))......))))))))))))))).) ( -39.90) >DroEre_CAF1 4320 98 + 1 UUUCU-------UUGCGUAGACCUCGGCGCCGUCUCCGAUUCCAGUUCACUGCUG--UUUUGGGUGCACAGCUGCCGGAAGCGGUAAUUGGAGUUGGAAUCGGCGUC ..(((-------......)))....(((((((..((((((((((((....(((((--((((.(((((...)).))).)))))))))))))))))))))..))))))) ( -44.90) >DroYak_CAF1 3753 103 + 1 UUUUUUUUUU--UCGCGUAGACCUCGGCGCCGUCUCCGAUUCCAGUUCACUGCUG--UUUUGGGUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGCGUC ..........--.............(((((((..(((((((((((((.....(..--((((.(((((...)).))).))))..).)))))))))))))..))))))) ( -40.30) >DroPer_CAF1 6865 95 + 1 CUCUCGAUUCAAUUGCG--AGCCUC--CGCUGUGUGUGUGUUCUGCUCUCUGCUC--U-----CUGCACAGUUGCCGGAAAUGGAAAUUAA-AUUGAAAACGAAGAC .(((((.(((((((...--..((((--(((((((((.(.(....((.....)).)--.-----))))))))....))))...))......)-))))))..)).))). ( -20.40) >consensus UUUUU____U__UCGCGUAGACCUCGGCGCCGUCUCCGAUUCCAGCUCACUGCUG__UUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUC ..............((((........)))).(.(((((((((((((((...((((.............))))..(((....)))......))))))))))))))).) (-24.07 = -24.67 + 0.60)

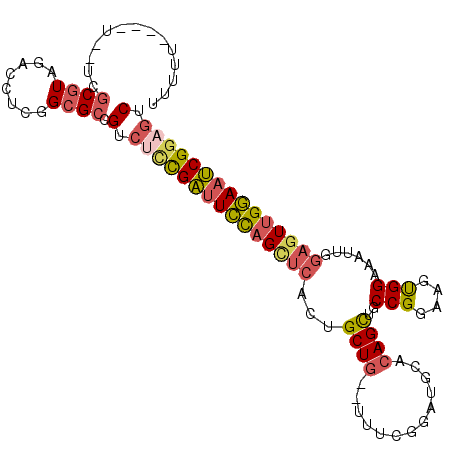
| Location | 18,475,048 – 18,475,141 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18475048 93 + 22224390 CGUCUCCGAUUCUAGCU---CAUCGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUCGAAA---UGUAAAUGAAGG---- ((.((((((((((((((---(...(((((((.......)))))))..(((....)))......))))))))))))))).))...---............---- ( -40.50) >DroSec_CAF1 3732 93 + 1 CGUCUUCGAUUCCAGCU---CACUGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUCGAAA---UGUAAAUGGAGA---- ((.((((((((((((((---(...(((((((.......)))))))..(((....)))......))))))))))))))).))...---............---- ( -39.60) >DroSim_CAF1 2870 93 + 1 CGUCUUCGAUUCCAGCU---CACUGCUGUGUUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUCGAAA---UGUAAAUAAUGA---- ((.((((((((((((((---(...(((((((.......)))))))..(((....)))......))))))))))))))).))...---............---- ( -39.60) >DroEre_CAF1 4343 91 + 1 CGUCUCCGAUUCCAGUU---CACUGCUG--UUUUGGGUGCACAGCUGCCGGAAGCGGUAAUUGGAGUUGGAAUCGGCGUCGAAA---UGUAAAUGAAGA---- ((.(.(((((((((..(---(..(((((--((((.(((((...)).))).)))))))))....))..))))))))).).))...---............---- ( -36.40) >DroYak_CAF1 3781 91 + 1 CGUCUCCGAUUCCAGUU---CACUGCUG--UUUUGGGUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGCGUCGAAA---UGUAAAUGAAGA---- ((.(.(((((((((..(---(....(..--((((.(((((...)).))).))))..)......))..))))))))).).))...---............---- ( -31.30) >DroAna_CAF1 3897 102 + 1 AGUUUUGGAAGCCGGAUUCGUACGACUGU-UUUUGGAAGUACAGCUGCCGGAAAUGGAAAUUGAAAUUGAAACGAAAAACGAAAAAACGAAAAAAAAGAGAUG .((((..(...((((....((((..((..-....))..)))).....)))).(((....)))....)..)))).............................. ( -16.40) >consensus CGUCUCCGAUUCCAGCU___CACUGCUGU_UUUCGGAUGCACAGCUGCCGGAAGUGGAAAUUGGAGUUGGAAUCGGAGUCGAAA___UGUAAAUGAAGA____ ((.((((((((((((((.....(((((....(((((..((...))..)))))))))).......)))))))))))))).))...................... (-23.20 = -23.18 + -0.02)


| Location | 18,475,048 – 18,475,141 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -15.38 |
| Energy contribution | -16.52 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18475048 93 - 22224390 ----CCUUCAUUUACA---UUUCGACUCCGAUUCCAACUCCAAUUUCCACUUCCGGCAGCUGUGCAUCCGAAACACAGCGAUG---AGCUAGAAUCGGAGACG ----............---...((.(((((((((...(((......((......))..((((((.........))))))...)---))...))))))))).)) ( -25.80) >DroSec_CAF1 3732 93 - 1 ----UCUCCAUUUACA---UUUCGACUCCGAUUCCAACUCCAAUUUCCACUUCCGGCAGCUGUGCAUCCGAAACACAGCAGUG---AGCUGGAAUCGAAGACG ----............---...((.((.((((((((.((((.....((......))..((((((.........)))))).).)---)).)))))))).)).)) ( -25.90) >DroSim_CAF1 2870 93 - 1 ----UCAUUAUUUACA---UUUCGACUCCGAUUCCAACUCCAAUUUCCACUUCCGGCAGCUGUGCAUCCGAAACACAGCAGUG---AGCUGGAAUCGAAGACG ----............---...((.((.((((((((.((((.....((......))..((((((.........)))))).).)---)).)))))))).)).)) ( -25.90) >DroEre_CAF1 4343 91 - 1 ----UCUUCAUUUACA---UUUCGACGCCGAUUCCAACUCCAAUUACCGCUUCCGGCAGCUGUGCACCCAAAA--CAGCAGUG---AACUGGAAUCGGAGACG ----............---...((.(.(((((((((..(((.....(((....)))..(((((.........)--)))).).)---)..))))))))).).)) ( -24.80) >DroYak_CAF1 3781 91 - 1 ----UCUUCAUUUACA---UUUCGACGCCGAUUCCAACUCCAAUUUCCACUUCCGGCAGCUGUGCACCCAAAA--CAGCAGUG---AACUGGAAUCGGAGACG ----............---...((.(.(((((((((..(((.....((......))..(((((.........)--)))).).)---)..))))))))).).)) ( -23.00) >DroAna_CAF1 3897 102 - 1 CAUCUCUUUUUUUUCGUUUUUUCGUUUUUCGUUUCAAUUUCAAUUUCCAUUUCCGGCAGCUGUACUUCCAAAA-ACAGUCGUACGAAUCCGGCUUCCAAAACU ...............(((((..((.....)).....................((((.(..(((((..(.....-...)..)))))..))))).....))))). ( -9.50) >consensus ____UCUUCAUUUACA___UUUCGACUCCGAUUCCAACUCCAAUUUCCACUUCCGGCAGCUGUGCAUCCAAAA_ACAGCAGUG___AGCUGGAAUCGAAGACG .........................(((((((((((..........((......))..(((((...........)))))..........)))))))))))... (-15.38 = -16.52 + 1.14)

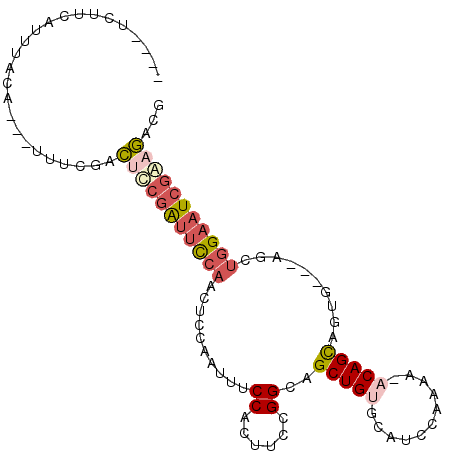

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:11 2006