
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,474,621 – 18,474,712 |
| Length | 91 |
| Max. P | 0.994029 |

| Location | 18,474,621 – 18,474,712 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.14 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -16.75 |
| Energy contribution | -18.18 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
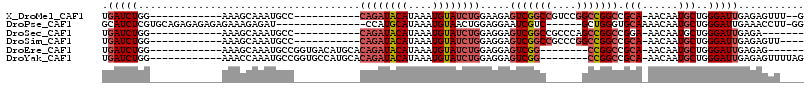
>X_DroMel_CAF1 18474621 91 + 22224390 UGAUCUGG------------AAAGCAAAUGCC-----------CAGAUACAUAAAUGUAUCUGGAAGAGUCGGCCGUCCGGCCGGCCGCA-AACAAUGCUGGGAUUGAGAGUUU--G .(((((..------------..((((..((((-----------((((((((....)))))))))..(.(((((((....)))))))))))-.....)))).)))))........--. ( -35.10) >DroPse_CAF1 6490 95 + 1 GCAUCUCGUGCAGAGAGAGAGAAAGAGAU---------------CCAUGCAUAAAUGUAACUGGAGGAAUCGUC------GCUGGGUGCAAAACAAUGCUGGGAUUGAAACCUU-GG ...((((.....))))..(((.....(((---------------((..((((...((((.(..(.((.....))------.)..).)))).....))))..))))).....)))-.. ( -19.60) >DroSec_CAF1 3321 86 + 1 UGAUCUGG------------AAAGCAAAUGCC-----------CAGAUACAUAAAUGUAUCUGGAGGAGUCGGCCGCCCAGCCGGCCGGA-AACAAUGCUGGGAUUGAGA------- .(((((..------------..((((.....(-----------((((((((....))))))))).(...(((((((......))))))).-..)..)))).)))))....------- ( -33.00) >DroSim_CAF1 2448 89 + 1 UGAUCUGG------------AAAGCAAAUGCC-----------CAGAUACAUAAAUGUAUCUGGAGGAGUCGGCCGCCCGGCCGGCCGCA-AACAAUGCUGGGAUUGAGAGUU---- .(((((..------------..((((..((((-----------((((((((....)))))))))..(.(((((((....)))))))))))-.....)))).))))).......---- ( -34.90) >DroEre_CAF1 3939 90 + 1 UGAUCUGG------------AAAGCAAAUGCCGGUGACAUGCACAGAUACAUAAAUGUAUCUGGAGGAGUCGG--------CCGGCCGCA-AACAAUGCUGGGAUUGAGAG------ .(((((.(------------.........(((((((((.....((((((((....)))))))).....))).)--------))))).(((-.....)))).))))).....------ ( -29.70) >DroYak_CAF1 3354 96 + 1 UGAUCUGG------------AAACCAAAUGCCGGUGCCAUGCACAGAUACAUAAAUGUAUCUGGAGGAGUCGG--------CCGGCCGCA-AACAAUGCUGGGAUUGAGAGUUUUAG ...(((.(------------(..(((...(((((((((...(.((((((((....)))))))))..).))..)--------))))).(((-.....))))))..)).)))....... ( -30.90) >consensus UGAUCUGG____________AAAGCAAAUGCC___________CAGAUACAUAAAUGUAUCUGGAGGAGUCGGC______GCCGGCCGCA_AACAAUGCUGGGAUUGAGAGUU____ .(((((.....................................((((((((....)))))))).....(((((((....))))))).(((......)))..)))))........... (-16.75 = -18.18 + 1.43)


| Location | 18,474,621 – 18,474,712 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.14 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -12.34 |
| Energy contribution | -13.91 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
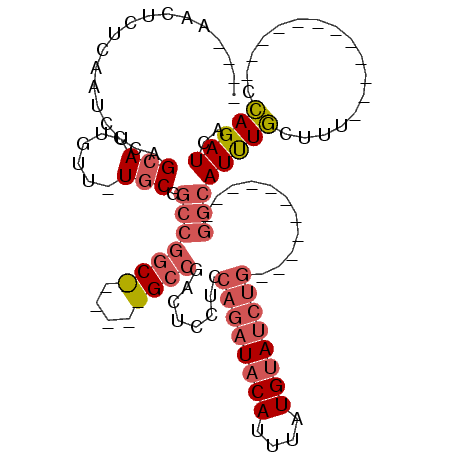
>X_DroMel_CAF1 18474621 91 - 22224390 C--AAACUCUCAAUCCCAGCAUUGUU-UGCGGCCGGCCGGACGGCCGACUCUUCCAGAUACAUUUAUGUAUCUG-----------GGCAUUUGCUUU------------CCAGAUCA .--..............((((.(((.-..((((((......))))))......(((((((((....))))))))-----------))))..))))..------------........ ( -30.60) >DroPse_CAF1 6490 95 - 1 CC-AAGGUUUCAAUCCCAGCAUUGUUUUGCACCCAGC------GACGAUUCCUCCAGUUACAUUUAUGCAUGG---------------AUCUCUUUCUCUCUCUCUGCACGAGAUGC ..-..((........)).(((((....((((...((.------((.((....((((((.........)).)))---------------)......)))).))...))))...))))) ( -13.90) >DroSec_CAF1 3321 86 - 1 -------UCUCAAUCCCAGCAUUGUU-UCCGGCCGGCUGGGCGGCCGACUCCUCCAGAUACAUUUAUGUAUCUG-----------GGCAUUUGCUUU------------CCAGAUCA -------..........((((.(((.-..((((((......))))))......(((((((((....))))))))-----------))))..))))..------------........ ( -29.70) >DroSim_CAF1 2448 89 - 1 ----AACUCUCAAUCCCAGCAUUGUU-UGCGGCCGGCCGGGCGGCCGACUCCUCCAGAUACAUUUAUGUAUCUG-----------GGCAUUUGCUUU------------CCAGAUCA ----.............((((.....-(((((.(((((....)))))...)).(((((((((....))))))))-----------))))..))))..------------........ ( -30.00) >DroEre_CAF1 3939 90 - 1 ------CUCUCAAUCCCAGCAUUGUU-UGCGGCCGG--------CCGACUCCUCCAGAUACAUUUAUGUAUCUGUGCAUGUCACCGGCAUUUGCUUU------------CCAGAUCA ------............(((.....-))).(((((--------..(((.....((((((((....)))))))).....))).)))))(((((....------------.))))).. ( -26.20) >DroYak_CAF1 3354 96 - 1 CUAAAACUCUCAAUCCCAGCAUUGUU-UGCGGCCGG--------CCGACUCCUCCAGAUACAUUUAUGUAUCUGUGCAUGGCACCGGCAUUUGGUUU------------CCAGAUCA .......(((..(..((((((.....-))).(((((--------..........((((((((....))))))))(((...))))))))...)))..)------------..)))... ( -26.10) >consensus ____AACUCUCAAUCCCAGCAUUGUU_UGCGGCCGGC______GCCGACUCCUCCAGAUACAUUUAUGUAUCUG___________GGCAUUUGCUUU____________CCAGAUCA ..................(((......))).(((((((....))))........((((((((....))))))))...........)))(((((.................))))).. (-12.34 = -13.91 + 1.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:08 2006