
| Sequence ID | X_DroMel_CAF1 |
|---|---|
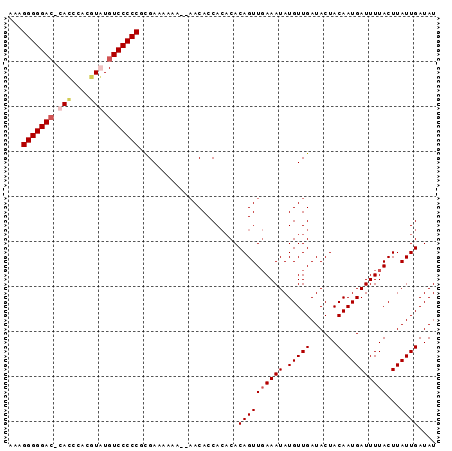
| Location | 18,469,608 – 18,469,740 |
| Length | 132 |
| Max. P | 0.960773 |

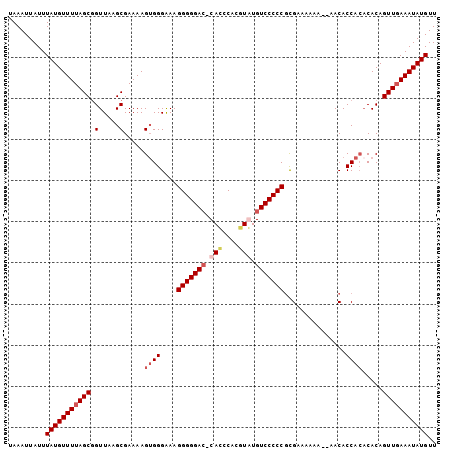
| Location | 18,469,608 – 18,469,702 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.72 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18469608 94 - 22224390 AAAGGGGGAC-CAUCCACGUAUGUCCCCCGUGAAAAAAAGAACACCACACACAGUUGAAAUAUGUUGAUACUACAAUGAUUUUACUUAUUGAUAU ...(((((((-...........)))))))(((.............)))...((((((((((.(((((......)))))))))))...)))).... ( -18.92) >DroSec_CAF1 13184 90 - 1 AAAGGGGGAC-CACCCACUUGUGUCCCCCGCGAAAAAA--AACACCA--CACAGUUUAAAUAUGUUGAUACUACAAUGAUUUUACUUAUUGAUAU ...(((((((-((......)).)))))))........(--(((....--....))))................((((((......)))))).... ( -19.40) >DroSim_CAF1 10025 94 - 1 AAAGGGGGAA-CACCCACGUGUGUCCCCCGCGAAAAAAAAAACACCACACACAGUUGAAAUAUGUUGAUACUACAAUGAUUUUACUUAUUGAUAU ...(((((((-(((....)))).))))))......................((((((((((.(((((......)))))))))))...)))).... ( -20.40) >DroEre_CAF1 10270 90 - 1 GAAGGGGGACCCACCCACGUAUGUCCCCCUCGAAAA-----ACACCACACACAGUUGAAAUAUGUUGAUACUACAAUGAUUUUACUUAUUGAUAU ((.(((((((..((....))..))))))))).....-----..........((((((((((.(((((......)))))))))))...)))).... ( -19.60) >consensus AAAGGGGGAC_CACCCACGUAUGUCCCCCGCGAAAAAA__AACACCACACACAGUUGAAAUAUGUUGAUACUACAAUGAUUUUACUUAUUGAUAU ...(((((((.(((....))).)))))))......................((((((((((.(((((......)))))))))))...)))).... (-17.60 = -18.72 + 1.12)


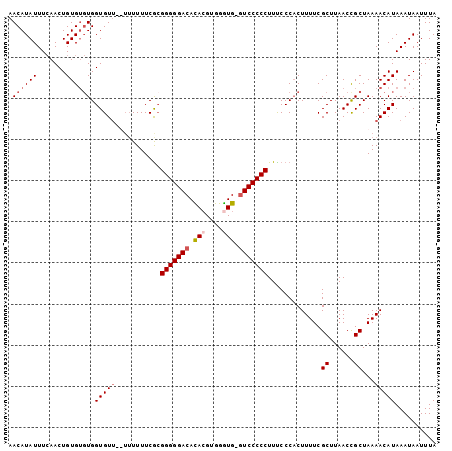
| Location | 18,469,637 – 18,469,740 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.75 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18469637 103 + 22224390 AACAUAUUUCAACUGUGUGUGGUGUUCUUUUUUUCACGGGGGACAUACGUGGAUG-GUCCCCCUUUCCCACUUUUCGCUUAACCGCUAAAACAUAAAUAAUUUA .............(((((((((((..........)))(((((((...........-))))))).....))).....((......))....)))))......... ( -21.50) >DroSec_CAF1 13213 99 + 1 AACAUAUUUAAACUGUG--UGGUGUU--UUUUUUCGCGGGGGACACAAGUGGGUG-GUCCCCCUUUCCCACUUUUCGCUUAACCGCUAAAACAUAAAUAAUUUA ..(((((.......)))--))(((((--((.....((((.((....(((((((.(-(....))...)))))))....))...)))).))))))).......... ( -25.90) >DroSim_CAF1 10054 103 + 1 AACAUAUUUCAACUGUGUGUGGUGUUUUUUUUUUCGCGGGGGACACACGUGGGUG-UUCCCCCUUUCCCACUUUUCGCUUAACCGCUAAAACAUAAAUAAUUUA .((((((.......)))))).(((((((.........((((((.((((....)))-))))))).............((......)).))))))).......... ( -25.30) >DroEre_CAF1 10299 99 + 1 AACAUAUUUCAACUGUGUGUGGUGU-----UUUUCGAGGGGGACAUACGUGGGUGGGUCCCCCUUCCCCCCUUUUCGCUUAACUGCUAAAACAUAAAUAAUUUA .((((((.......)))))).((((-----(((..(((((((((.(((....))).)))))))))...........((......)).))))))).......... ( -28.40) >consensus AACAUAUUUCAACUGUGUGUGGUGUU__UUUUUUCGCGGGGGACACACGUGGGUG_GUCCCCCUUUCCCACUUUUCGCUUAACCGCUAAAACAUAAAUAAUUUA .((((((.......)))))).(((((...........(((((((.(((....))).))))))).............((......))...))))).......... (-19.50 = -20.75 + 1.25)

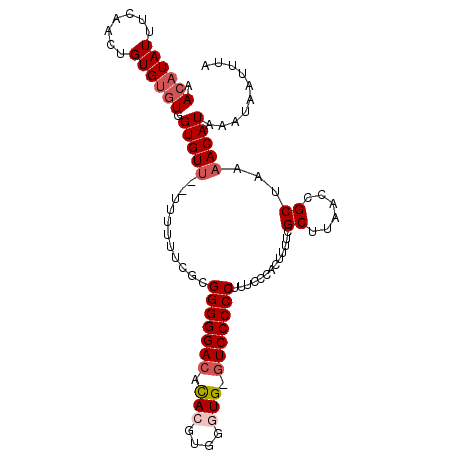
| Location | 18,469,637 – 18,469,740 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.61 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18469637 103 - 22224390 UAAAUUAUUUAUGUUUUAGCGGUUAAGCGAAAAGUGGGAAAGGGGGAC-CAUCCACGUAUGUCCCCCGUGAAAAAAAGAACACCACACACAGUUGAAAUAUGUU .........(((((((((((.(.....).....((((....(((((((-...........)))))))((..........)).)))).....))))))))))).. ( -27.30) >DroSec_CAF1 13213 99 - 1 UAAAUUAUUUAUGUUUUAGCGGUUAAGCGAAAAGUGGGAAAGGGGGAC-CACCCACUUGUGUCCCCCGCGAAAAAA--AACACCA--CACAGUUUAAAUAUGUU ((((((.....((((((.((((....(((..(((((((...((....)-).))))))).)))...)))).....))--))))...--...))))))........ ( -25.00) >DroSim_CAF1 10054 103 - 1 UAAAUUAUUUAUGUUUUAGCGGUUAAGCGAAAAGUGGGAAAGGGGGAA-CACCCACGUGUGUCCCCCGCGAAAAAAAAAACACCACACACAGUUGAAAUAUGUU .........(((((((((((.(.....).....((((....(((((((-(((....)))).))))))...............)))).....))))))))))).. ( -27.61) >DroEre_CAF1 10299 99 - 1 UAAAUUAUUUAUGUUUUAGCAGUUAAGCGAAAAGGGGGGAAGGGGGACCCACCCACGUAUGUCCCCCUCGAAAA-----ACACCACACACAGUUGAAAUAUGUU .........(((((((((((.(.....).....(((((((.((((......))).).....)))))))......-----............))))))))))).. ( -26.00) >consensus UAAAUUAUUUAUGUUUUAGCGGUUAAGCGAAAAGUGGGAAAGGGGGAC_CACCCACGUAUGUCCCCCGCGAAAAAA__AACACCACACACAGUUGAAAUAUGUU .........(((((((((((.(.....).....((((....(((((((.(((....))).)))))))...............)))).....))))))))))).. (-21.99 = -23.61 + 1.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:38:06 2006