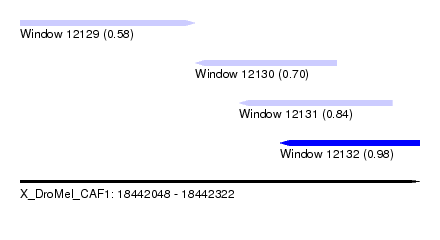
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,442,048 – 18,442,322 |
| Length | 274 |
| Max. P | 0.980218 |

| Location | 18,442,048 – 18,442,168 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.17 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18442048 120 + 22224390 UCGUGGAUCGGGAGGUCUGUUUAUUUGCUGUUGCAUCUCGGCGCUCCCACUGCUCAUGUUCACUCCUACAGGUCCGCCAUAUGGCGGAAUCUUGUUCGUGUCCGGGUUGCUUCCACUCUG ..(((((..((((((.(((......(((....)))...)))).)))))...((...((..(((....(((((((((((....))))))..)))))..)))..))....)).))))).... ( -42.70) >DroSec_CAF1 8560 117 + 1 GCGAGGAUCGGGAGGUCUCCUGAUUUGCU---GCAUCUCGGCGCUCCCAUUGUUCAAGUUCACUCCUCCAGGUCCGCCAUAUGGCGGAAUCUUGUUCGUGUCCGGGUUGCUUCCAUUCUG ((((..(((((((....))))))))))).---(((.((((((((.......(..((((......((....))((((((....))))))..))))..)))).))))).))).......... ( -39.01) >DroEre_CAF1 6714 111 + 1 GCGAGGAUCGGGAGCGAUCCUGACGUGCUGCUGCGUCUCGGCGCUGCCAUUGCAUACGUUGAU---------UCUGCCAGCUGGCACACUCUUGCUCAUGUCCGGGUUGCUUCCACUCUG ..(((....(((((((((((.((((((..((...(..((((((.(((....)))..)))))).---------.)((((....)))).......)).)))))).))))))))))).))).. ( -44.30) >consensus GCGAGGAUCGGGAGGUCUCCUGAUUUGCUG_UGCAUCUCGGCGCUCCCAUUGCUCAAGUUCACUCCU_CAGGUCCGCCAUAUGGCGGAAUCUUGUUCGUGUCCGGGUUGCUUCCACUCUG ....(((((((((....)))))).........(((.((((((((.......))...................((((((....))))))...........).))))).))).)))...... (-28.27 = -28.17 + -0.10)



| Location | 18,442,168 – 18,442,265 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -18.53 |
| Energy contribution | -17.10 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18442168 97 - 22224390 UCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC------------CAUGAACAUGU-CACCACUUAACCUUGUGAGUCCUCGACGCAAUG----------UCAAUACCUAUCAC .....((((((((((((......)))))(((((....))------------)))......))-)(((((........))).))...))))......----------.............. ( -25.10) >DroSec_CAF1 8677 109 - 1 UCCAAGUCGGAUGGAGCAGCCUAACUCCAUGGGUAUGCC-----------CCAUGAAUAUGCCCACCACUCAACCUUAUGAGUCCUCGGCGCAAUGUAAUACCCUAUCAAUACCUAUCAC ........((((((((........))))))((((((...-----------.(((.....((((....(((((......)))))....))))..)))..))))))........))...... ( -26.70) >DroYak_CAF1 5237 106 - 1 UCCAACUCGGAUGGAGCAGCCCGG---CAUGGGCAUGCAGCAGGGCAUGGCCAUGAACAUGC-CACCGCCCAAUCACAUGAGUCCGCGGCACAAUG----------UCAAUCCCUAUCAU .........(((((.(((((((..---...)))).))).(((.((((((........)))))-).((((....((....))....)))).....))----------)......))))).. ( -34.00) >consensus UCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC___________CCAUGAACAUGC_CACCACUCAACCUUAUGAGUCCUCGGCGCAAUG__________UCAAUACCUAUCAC .........(((((.((((((((......))))).))).....................(((.....(((((......))))).....)))......................))))).. (-18.53 = -17.10 + -1.43)

| Location | 18,442,198 – 18,442,303 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -24.69 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18442198 105 - 22224390 GG-AGUCGGAUUGCUUCCGUUGCCUUUGCAGAUGCUCA-GUCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC------------CAUGAACAUGU-CACCACUUAACCUUGUG ((-((((((..((((((..((((....)))).......-((((.....))))))))))..))))))))(((((....))------------)))........-...(((........))) ( -36.40) >DroSec_CAF1 8717 108 - 1 GGCGGUCGAAUUGCU-CCGUUGCCUUUGCAAAUGCUCAAAUCCAAGUCGGAUGGAGCAGCCUAACUCCAUGGGUAUGCC-----------CCAUGAAUAUGCCCACCACUCAACCUUAUG ((((((.....((((-((.((((....))))........((((.....)))))))))))))......((((((.....)-----------))))).....)))................. ( -31.30) >DroYak_CAF1 5267 114 - 1 GG-CGUCGGACUGCUGCCGUUGCCGCUGCAGAUGCUCA-GUCCAACUCGGAUGGAGCAGCCCGG---CAUGGGCAUGCAGCAGGGCAUGGCCAUGAACAUGC-CACCGCCCAAUCACAUG ((-((.(((.(....)))).))))((((((..(((((.-((((.....)))).)))))((((..---...)))).)))))).((((.((((.((....))))-))..))))......... ( -54.60) >consensus GG_AGUCGGAUUGCU_CCGUUGCCUUUGCAGAUGCUCA_GUCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC___________CCAUGAACAUGC_CACCACUCAACCUUAUG ((.((((((.(((((.((.((((....))))........((((.....))))))))))).)))))).((((..((((..............))))..))))....))............. (-24.69 = -24.37 + -0.32)


| Location | 18,442,226 – 18,442,322 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -44.67 |
| Consensus MFE | -29.17 |
| Energy contribution | -28.29 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18442226 96 - 22224390 ---UCAC---------AUGGGCAGG------GGCAUGGG-AGUCGGAUUGCUUCCGUUGCCUUUGCAGAUGCUCA-GUCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC- ---...(---------(((((((((------((((((((-(((......))))))).)))))))))((.(((((.-((((.....)))).)))))..))....)))))........- ( -43.80) >DroSec_CAF1 8747 105 - 1 ---UCACAUGA-CCACAUGGGCAGG------GGCAUGGGCGGUCGAAUUGCU-CCGUUGCCUUUGCAAAUGCUCAAAUCCAAGUCGGAUGGAGCAGCCUAACUCCAUGGGUAUGCC- ---...((((.-((.((((((((((------((((((((((((...))))).-))).)))))))))...(((((..((((.....)))).)))))........)))))))))))..- ( -44.30) >DroYak_CAF1 5306 112 - 1 CCACCACAUGAACCACAUGGGCAGGGGCAUGGGCAUGGG-CGUCGGACUGCUGCCGUUGCCGCUGCAGAUGCUCA-GUCCAACUCGGAUGGAGCAGCCCGG---CAUGGGCAUGCAG (((((.((((..((.........))..))))))..))).-.......((((((((..(((((..((...(((((.-((((.....)))).))))))).)))---))..)))).)))) ( -45.90) >consensus ___UCACAUGA_CCACAUGGGCAGG______GGCAUGGG_AGUCGGAUUGCU_CCGUUGCCUUUGCAGAUGCUCA_GUCCAAGUCGGAUGGAGCAGCCUGGCUCCAUGGGUAUGCC_ ...((............((((((........((((..((.(((......))).))..))))........)))))).((((.....)))))).((((((((......))))).))).. (-29.17 = -28.29 + -0.88)

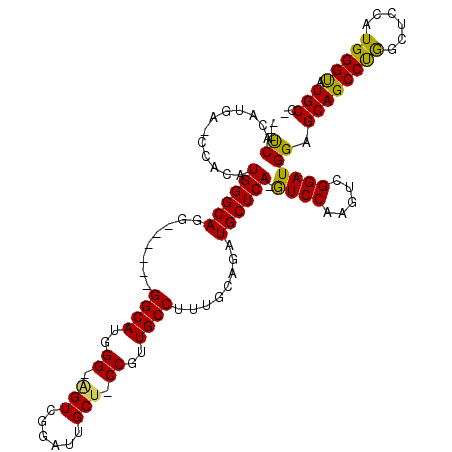

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:58 2006