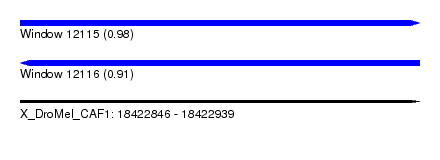
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,422,846 – 18,422,939 |
| Length | 93 |
| Max. P | 0.984775 |

| Location | 18,422,846 – 18,422,939 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18422846 93 + 22224390 UUCCUGGCACACCUUCACCCCCACCUCCAUCUUCACCAGCCCCCACUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU ....((((............(((..(((((.(((....(((........))).(....)))).)))))..)))....(....)))))...... ( -25.10) >DroSec_CAF1 45775 90 + 1 UUCCUGGCUCACCUCCAGCCCCACCACCAACUUCACCU---CCAACUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU ((((.((((.......)))).................(---(((..((((........))))..))))...))))..(((...)))....... ( -20.80) >DroSim_CAF1 46029 90 + 1 UUCCUGGCUCACCUCCACCCCCACCACCAACUUUACCA---CCAACUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU ....((((...........................(((---(((..((((........))))..)))...)))....(....)))))...... ( -18.20) >DroEre_CAF1 47357 78 + 1 CCUCCAGUUC------------ACCUCCACCUCCAUAU---CCACCUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU .....((((.------------...((((..(((((.(---((...(((....)))...))).)))))..))))...(((...)))...)))) ( -21.60) >DroYak_CAF1 27460 90 + 1 CUCCUGGUUCAACUCCAACUCCAACUCCAACUCCAUAU---CCAGCUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU ....((((.................((((..(((((.(---((.((....)).(....)))).)))))..))))...(....)))))...... ( -23.80) >consensus UUCCUGGCUCACCUCCACCCCCACCUCCAACUUCACCU___CCAACUCCGGCAGGAAACGGAAAUGGAAAUGGAAACGGCAACGCCAUCAACU ....((((.................................(((..((((........))))..)))....(....)(....)))))...... (-14.38 = -14.74 + 0.36)


| Location | 18,422,846 – 18,422,939 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -18.08 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18422846 93 - 22224390 AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGUGGGGGCUGGUGAAGAUGGAGGUGGGGGUGAAGGUGUGCCAGGAA ......((((...(((.....(((..((((((((((((((.(((........))).))...))))))))))))..)))..)))..)))).... ( -36.80) >DroSec_CAF1 45775 90 - 1 AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGUUGG---AGGUGAAGUUGGUGGUGGGGCUGGAGGUGAGCCAGGAA ......((((.(..((...((((.((((((..(((...(((.(((....)))---)))......)))..)))))).))))))..))))).... ( -30.60) >DroSim_CAF1 46029 90 - 1 AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGUUGG---UGGUAAAGUUGGUGGUGGGGGUGGAGGUGAGCCAGGAA ......((((.(..((...(((((..((((..((((((.(..(((....)))---..).)))..)))..))))..)))))))..))))).... ( -35.70) >DroEre_CAF1 47357 78 - 1 AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGGUGG---AUAUGGAGGUGGAGGU------------GAACUGGAGG ((((.(((((...)))..((((((((((((.((((..(((....)))..)))---).))))))))))))))------------.))))..... ( -34.40) >DroYak_CAF1 27460 90 - 1 AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGCUGG---AUAUGGAGUUGGAGUUGGAGUUGGAGUUGAACCAGGAG ......(((((...(((.(((((.(((((.((((((.(((.((....)).))---).)))))).))))).))))).)))...))..))).... ( -31.80) >consensus AGUUGAUGGCGUUGCCGUUUCCAUUUCCAUUUCCGUUUCCUGCCGGAGUUGG___AGGUGAAGUUGGAGGUGGGGGUGGAGGUGAGCCAGGAA ......((((....(((..((((((((((((((((.((((....)))).)))........))).))))))))))..)))......)))).... (-18.08 = -20.00 + 1.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:45 2006