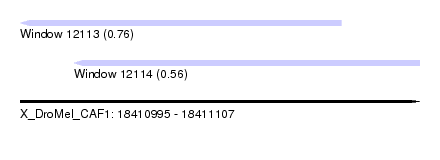
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,410,995 – 18,411,107 |
| Length | 112 |
| Max. P | 0.763008 |

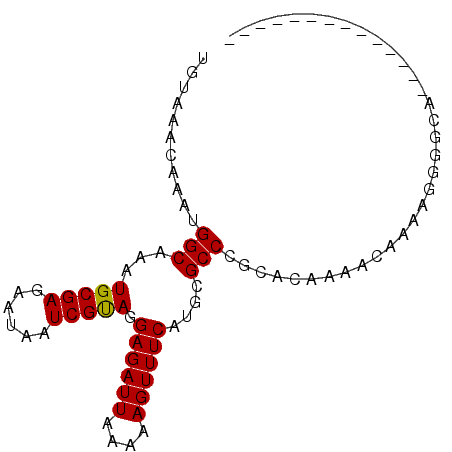
| Location | 18,410,995 – 18,411,085 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -12.42 |
| Energy contribution | -12.28 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18410995 90 - 22224390 UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUGCGCCCGCACAAAGCAAAAGGGGCAGCUCGAGCCCCAG- ...........(((......((......))((((((((((....))))))))))))).((.....))....(((((.......)))))..- ( -29.20) >DroGri_CAF1 37054 78 - 1 UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACAGCACAAUAAGAUCG-A------------ (((........(((...(((((......))))).((((((....))))))....))).((...))))).........-.------------ ( -12.60) >DroSec_CAF1 33877 90 - 1 UGUAAGCAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUGCGCCCGCACAAAGCAAAAGGGGCAGCCCGAGCCGCAG- ...........(((....((..........((((((((((....))))))))))((((((.....)).....)))).))....)))....- ( -26.20) >DroEre_CAF1 36219 77 - 1 UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGCAGGAGAUUAAAAAGUUUCCUGCGCCCGCACA-AGCAAAAGGGGCA-------------G ............((....))((......))((((((((((....))))))))))((((((...-.)).....)))).-------------. ( -22.80) >DroMoj_CAF1 11217 77 - 1 UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACUGUACGACGGGAGCG-------------- ...........(((...(((((......))))).((((((....))))))....)))(((.(((.....)))..)))-------------- ( -17.50) >DroAna_CAF1 11670 90 - 1 UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACAAAAGCAGGGGGUAAAAGCAGGACGCAG- .................((((....(((((......)))))....((((..(((.(((((......)).))).))).))))....)))).- ( -20.80) >consensus UGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACAAAACAAAAGGGGCA______________ ...........(((...(((((......))))).((((((....))))))....))).................................. (-12.42 = -12.28 + -0.14)

| Location | 18,411,010 – 18,411,107 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -15.89 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18411010 97 - 22224390 CUCCUCCUCC-ACCAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCCUGCGCCCGCACAAAGCAAAAGGGG (((((.((((-.((((.(((((....))))).))))....(((((......)))))))))........((((..(((....)))..))))...))))) ( -27.60) >DroVir_CAF1 31965 97 - 1 CCCCGUUCCA-UUCGUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGUGCCCGCACAGCACAACGGAGC ....(((((.-((.((((((((((....))))))((((..(((((......))))).((((((....))))))...))))......)))))).))))) ( -26.60) >DroGri_CAF1 37057 97 - 1 ACCCUUUGCC-UUCAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACAGCACAAUAAGAU .........(-((..((((((.((((........(((...(((((......))))).((((((....))))))....))).))))))))))..))).. ( -21.40) >DroEre_CAF1 36222 96 - 1 CUCCUGCUCC-GCCAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGCAGGAGAUUAAAAAGUUUCCUGCGCCCGCACA-AGCAAAAGGGG ((((((((..-(((((.(((((....))))).)))))...((((.((....))((((((((((....))))))))))...))))..-)))...))))) ( -35.10) >DroWil_CAF1 30465 90 - 1 CCCCCUCUCUAUUCAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAGG---CCGCAUAUAC-AUAG---- ........((((..((((((((((....))))..(((...(((((......))))).((((((....))))))..)---))))))))..-))))---- ( -19.70) >DroMoj_CAF1 11219 93 - 1 CCA-GCU----CUUAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACUGUACGACGGGAG ((.-..(----(.(((((((((((....))))..(((...(((((......))))).((((((....))))))....))).)))).))).))..)).. ( -20.10) >consensus CCCCUUCUCC_UUCAUGUGUUUGUGUAAACAAAUGGCAAAUGCGAGAAUAAUCGUAGGAGAUUAAAAAGUUUCAUGCGCCCGCACAGAACAAAAGGGG ...............(((((((((....))))..(((...(((((......))))).((((((....))))))....))).)))))............ (-15.89 = -16.12 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:43 2006