
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,387,794 – 18,387,905 |
| Length | 111 |
| Max. P | 0.878849 |

| Location | 18,387,794 – 18,387,905 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -21.13 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
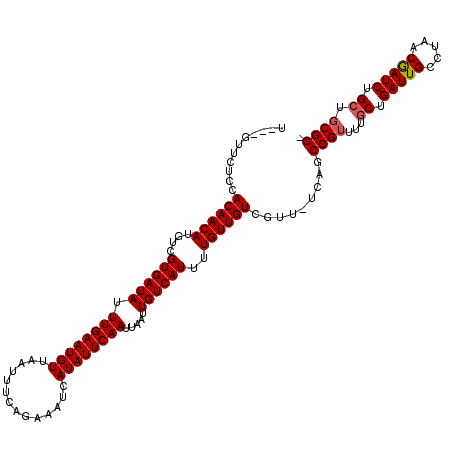
>X_DroMel_CAF1 18387794 111 + 22224390 --------UCCACAACAUGUCGUGAUAUUUGAAUGUUAAUUUCAAAAAUCAUAUUCAAUUAAUUGUCAUUUUGUUGUCGUU-UCAGCCGUUUUGCUGAUUGCGUAACGAUCUGCUGCGGC --------...((((((....((((((.((((((((..............)))))))).....))))))..))))))....-...(((((...((.(((((.....))))).)).))))) ( -27.54) >DroSim_CAF1 14137 116 + 1 U---GUUCUCCACAACAUGUCGUGAUAUUUGAAUGUUAAUUUCAGAAAUCAUAUUCAAUUAAUUGUCAUUUUGUUGUCGUUUUCAGCCGUUUUGCUGAUUGCCUAACGAUCUGCUGCGG- .---.....((.((.((.(((((((((...(((((.(((((...(((......)))....))))).)))))...))))((..(((((......)))))..))...))))).)).)).))- ( -25.90) >DroYak_CAF1 31605 115 + 1 UAUGGUUCUCCACAACAUGUCGUGAUAUUUGAAUGUUCUUUUCAGAAAUCAUAUUCAAUUAAUUGUCAUUUUGUUGUCGUU-UCAGCCGUUUUGCUGAUUGCCCAACAAUCUG---CGG- .((((((..(.((((((....((((((.(((((((((((....)))....)))))))).....))))))..)))))).)..-..))))))..(((.(((((.....))))).)---)).- ( -26.10) >consensus U___GUUCUCCACAACAUGUCGUGAUAUUUGAAUGUUAAUUUCAGAAAUCAUAUUCAAUUAAUUGUCAUUUUGUUGUCGUU_UCAGCCGUUUUGCUGAUUGCCUAACGAUCUGCUGCGG_ ...........((((((....((((((.((((((((..............)))))))).....))))))..)))))).........((((...((.(((((.....))))).)).)))). (-21.13 = -21.57 + 0.45)

| Location | 18,387,794 – 18,387,905 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18387794 111 - 22224390 GCCGCAGCAGAUCGUUACGCAAUCAGCAAAACGGCUGA-AACGACAACAAAAUGACAAUUAAUUGAAUAUGAUUUUUGAAAUUAACAUUCAAAUAUCACGACAUGUUGUGGA-------- .((((((((..((((..((...(((((......)))))-..))...........................(((.((((((.......)))))).)))))))..)))))))).-------- ( -29.10) >DroSim_CAF1 14137 116 - 1 -CCGCAGCAGAUCGUUAGGCAAUCAGCAAAACGGCUGAAAACGACAACAAAAUGACAAUUAAUUGAAUAUGAUUUCUGAAAUUAACAUUCAAAUAUCACGACAUGUUGUGGAGAAC---A -((((((((..((((.......(((((......)))))........................((((((.(((((.....)))))..)))))).....))))..)))))))).....---. ( -26.50) >DroYak_CAF1 31605 115 - 1 -CCG---CAGAUUGUUGGGCAAUCAGCAAAACGGCUGA-AACGACAACAAAAUGACAAUUAAUUGAAUAUGAUUUCUGAAAAGAACAUUCAAAUAUCACGACAUGUUGUGGAGAACCAUA -(((---(((..(((((.....(((((......)))))-.......................((((((.....((((....)))).))))))......)))))..))))))......... ( -24.90) >consensus _CCGCAGCAGAUCGUUAGGCAAUCAGCAAAACGGCUGA_AACGACAACAAAAUGACAAUUAAUUGAAUAUGAUUUCUGAAAUUAACAUUCAAAUAUCACGACAUGUUGUGGAGAAC___A .((((((((..((((.......(((((......)))))................................(((...((((.......))))...)))))))..))))))))......... (-20.99 = -21.77 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:37 2006