
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,376,869 – 18,376,961 |
| Length | 92 |
| Max. P | 0.755709 |

| Location | 18,376,869 – 18,376,961 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.55 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -10.23 |
| Energy contribution | -9.91 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
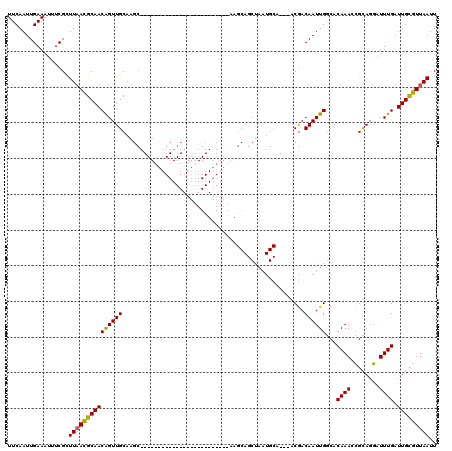
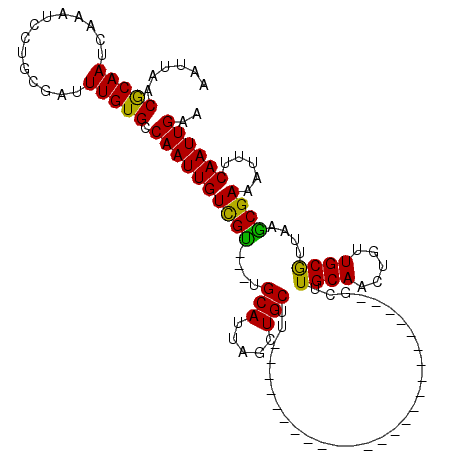
>X_DroMel_CAF1 18376869 92 + 22224390 UUCAAUUGAAAAUUCGCUUAACGCAACAGUUGCAAGC-------------------------AAGCAGCUAAUGCA---ACGACAAUUGGCACAAAUCGCAGGUUUUGAUUGCGUUAAUU .................((((((((((((((((((((-------------------------.....)))..))))---))...(((..((.......))..))).)).))))))))).. ( -22.10) >DroAna_CAF1 25454 75 + 1 UUCAAUUGAAAUUUCGUUUUAUGCAACAGUUGCA---------------------------------------------ACAACAAUUGCCACAAACUGUACAAUUUGAUUGCGUUAAUU ..((((..((.....((.....)).(((((((((---------------------------------------------(......))))....))))))....))..))))........ ( -11.10) >DroYak_CAF1 20439 120 + 1 UUCAAUUGAAAUUUCGCUUAACGCAACGGUUGCAAGCGGCUAAAGCAGCAAAAAGCUACAAAAAGCAGCUAAUGCACUAGCGACAAUUGGCACAAAGCGCAGGAUUUGAUUGUGUUAAUU .(((((((.....(((((....(((..((((((..((.......))(((.....))).......))))))..)))...)))))))))))).....(((((((.......))))))).... ( -31.30) >consensus UUCAAUUGAAAUUUCGCUUAACGCAACAGUUGCAAGC_________________________AAGCAGCUAAUGCA___ACGACAAUUGGCACAAACCGCAGGAUUUGAUUGCGUUAAUU .................(((((((((((((((...................................................))))))...((((........)))).))))))))).. (-10.23 = -9.91 + -0.33)


| Location | 18,376,869 – 18,376,961 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.55 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -10.79 |
| Energy contribution | -9.94 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18376869 92 - 22224390 AAUUAACGCAAUCAAAACCUGCGAUUUGUGCCAAUUGUCGU---UGCAUUAGCUGCUU-------------------------GCUUGCAACUGUUGCGUUAAGCGAAUUUUCAAUUGAA ..((((((((((........((((((......)))))).((---((((..(((.....-------------------------))))))))).))))))))))................. ( -23.30) >DroAna_CAF1 25454 75 - 1 AAUUAACGCAAUCAAAUUGUACAGUUUGUGGCAAUUGUUGU---------------------------------------------UGCAACUGUUGCAUAAAACGAAAUUUCAAUUGAA ...........((((..(((((((((.(..(((.....)))---------------------------------------------..)))))).))))......((....))..)))). ( -14.20) >DroYak_CAF1 20439 120 - 1 AAUUAACACAAUCAAAUCCUGCGCUUUGUGCCAAUUGUCGCUAGUGCAUUAGCUGCUUUUUGUAGCUUUUUGCUGCUUUAGCCGCUUGCAACCGUUGCGUUAAGCGAAAUUUCAAUUGAA ........((((..((.....(((((..(((.(((.((.((.((((((..((((((.....))))))...))).((....)).))).)).)).))))))..)))))....))..)))).. ( -30.80) >consensus AAUUAACGCAAUCAAAUCCUGCGAUUUGUGCCAAUUGUCGU___UGCAUUAGCUGCUU_________________________GCUUGCAACUGUUGCGUUAAGCGAAAUUUCAAUUGAA ......(((((..............))))).((((((((((....(((.....)))..............................((((.....))))....)))).....)))))).. (-10.79 = -9.94 + -0.85)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:31 2006