
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,376,439 – 18,376,598 |
| Length | 159 |
| Max. P | 0.981680 |

| Location | 18,376,439 – 18,376,558 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.67 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
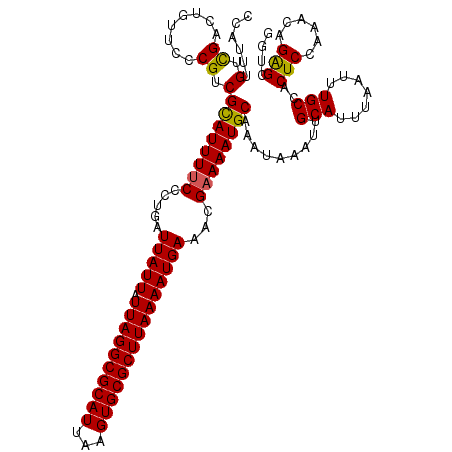
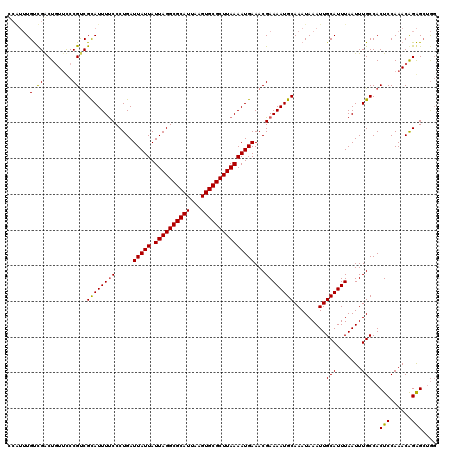
>X_DroMel_CAF1 18376439 119 + 22224390 CCC-AUGCCGGCUGUUCCCGUCGCAUUUUCCCUGAUUAUUAUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGGGAAAG ...-(((((((......)))..))))((((((((.(((((.((((((((((...))))))))))))))).....((((((((.....))))))))................)))))))). ( -37.20) >DroAna_CAF1 25018 118 + 1 UGAUUUGUUGACUUU-UCCGCCGUAUUUCCCCUGAUUAUUAUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGAGCUG- ...((((((......-...((..............(((((.((((((((((...))))))))))))))).....((((((((.....)))))))).....)).......))))))....- ( -23.69) >DroYak_CAF1 19993 120 + 1 CCGUUUGUCGACUGUUCCCGUCGCAUUUUCCCUGAUUAUUAUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACGGAGCUGG (((((((..(((.......)))((((((((.....(((((.((((((((((...)))))))))))))))...)))))))).........(((.......))).....)))))))...... ( -34.90) >consensus CCAUUUGUCGACUGUUCCCGUCGCAUUUUCCCUGAUUAUUAUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGAGCUGG ......(.((........)).)((((((((.....(((((.((((((((((...)))))))))))))))...)))))))).........(((.......)))..(((......))).... (-23.99 = -23.67 + -0.33)

| Location | 18,376,439 – 18,376,558 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -27.33 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
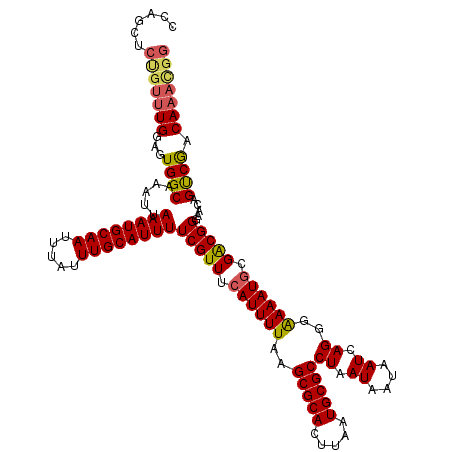
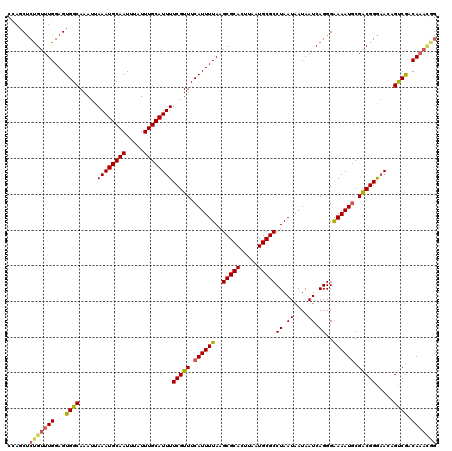
>X_DroMel_CAF1 18376439 119 - 22224390 CUUUCCCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAUAAUAAUCAGGGAAAAUGCGACGGGAACAGCCGGCAU-GGG .((((((((.(((((((((.((...((((((((.....))))))))...)))))))))))(((((.....)))))...........))))))))((((..(((......)))))))-... ( -37.80) >DroAna_CAF1 25018 118 - 1 -CAGCUCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAUAAUAAUCAGGGGAAAUACGGCGGA-AAAGUCAACAAAUCA -...(((((.(((((((((.((...((((((((.....))))))))...)))))))))))(((((.....)))))...........))))).......(((...-...)))......... ( -28.50) >DroYak_CAF1 19993 120 - 1 CCAGCUCCGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAUAAUAAUCAGGGAAAAUGCGACGGGAACAGUCGACAAACGG ......(((((((...((((........(((((.....)))))((..((((.((((((..(((((.....)))))((.((....)).))..)))))).))))..))..)))).))))))) ( -34.00) >consensus CCAGCUCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAUAAUAAUCAGGGAAAAUGCGACGGGAACAGUCGACAAACGG ......(((((((...((((.....((((((((.....))))))))(((((.((((((..(((((.....)))))((.((....)).))..)))))).))))).....)))).))))))) (-27.33 = -28.23 + 0.90)

| Location | 18,376,478 – 18,376,598 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -26.38 |
| Energy contribution | -27.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18376478 120 + 22224390 AUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGGGAAAGGGGAAGGGGGCUGCGCGGAGAGCUGUGCUGCCAUUUUAAU .((((((((((...))))))))))..........((((((((.....))))))))..(((.((..(((......)))..)).)))...(((.((((((....)))))).)))........ ( -40.00) >DroAna_CAF1 25057 105 + 1 AUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGAGCUG---------------GUGGGGAGCUGUGCUGCCAUUUUAAU .((((((((((...))))))))))..........((((((((.....)))))))).....(((((((......))).))---------------)).((.(((...))).))........ ( -31.20) >DroYak_CAF1 20033 120 + 1 AUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACGGAGCUGGGGAAAGGGGGCUGUGCGGAGAGCUGUGCUGCCAUUUUAAU .((((((((((...))))))))))..........((((((((.....))))))))..(((.(((((((....)))).))).)))....(((.(..(((....)))..).)))........ ( -42.00) >consensus AUUAGGCGCAUUAAGUGCGCUUAAAAUGAAACGAAAAUGCAAAUAAAUUGCAUUUAAUUUGCCACUCCAAACAGAGCUGGGG_AAGGGGGCUG_GCGGAGAGCUGUGCUGCCAUUUUAAU .((((((((((...))))))))))..........((((((((.....)))))))).........(((......)))............(((.((((((....)))))).)))........ (-26.38 = -27.50 + 1.12)



| Location | 18,376,478 – 18,376,598 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -27.78 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18376478 120 - 22224390 AUUAAAAUGGCAGCACAGCUCUCCGCGCAGCCCCCUUCCCCUUUCCCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAU .(((((((((.(((...((((((((.((((................)))).))))).))).....((((((((.....))))))))..))))))))))))(((((.....)))))..... ( -34.69) >DroAna_CAF1 25057 105 - 1 AUUAAAAUGGCAGCACAGCUCCCCAC---------------CAGCUCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAU .(((((((((..((...(((((..((---------------.......))..))))).)).....((((((((.....)))))))).....)))))))))(((((.....)))))..... ( -28.70) >DroYak_CAF1 20033 120 - 1 AUUAAAAUGGCAGCACAGCUCUCCGCACAGCCCCCUUUCCCCAGCUCCGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAU .(((((((((.(((...((((((((.(((((............)))..)).))))).))).....((((((((.....))))))))..))))))))))))(((((.....)))))..... ( -33.00) >consensus AUUAAAAUGGCAGCACAGCUCUCCGC_CAGCCCCCUU_CCCCAGCUCUGUUUGGAGUGGCAAAUUAAAUGCAAUUUAUUUGCAUUUUCGUUUCAUUUUAAGCGCACUUAAUGCGCCUAAU .(((((((((.(((...((((((((.((....................)).))))).))).....((((((((.....))))))))..))))))))))))(((((.....)))))..... (-27.78 = -28.45 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:29 2006