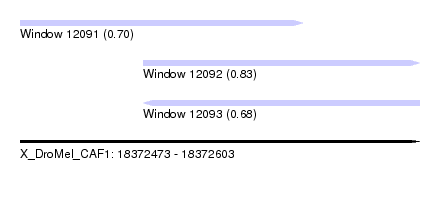
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,372,473 – 18,372,603 |
| Length | 130 |
| Max. P | 0.827130 |

| Location | 18,372,473 – 18,372,565 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18372473 92 + 22224390 GAGACAGAACGCUCUGGGCCAGUUUUGUGGGCAGUUUGCGGCCAGGCAAGCGA------AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAU ((((((((((((.....))..)))))))((.((((((...(((((((......------.)))))))))))))...))...........)))...... ( -27.30) >DroSec_CAF1 17212 92 + 1 GAGACAGAACGCUCUGGGCCAGUUUUGUGGGCAGUUUGCGGACAGGCAAGCGA------AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAU ((((((((((((.....))..)))))))((.((((((((...(((((......------.)))))))))))))...))...........)))...... ( -26.60) >DroYak_CAF1 16039 98 + 1 GAGACAGAACGCUCUGGGCCAGUUUUGUGGGCAGUUUGCGGCCAGGCAAGCGAAGCCGAAGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAU ((((((((((((.....))..)))))))((.((((((...(((((((...((....))..)))))))))))))...))...........)))...... ( -28.80) >consensus GAGACAGAACGCUCUGGGCCAGUUUUGUGGGCAGUUUGCGGCCAGGCAAGCGA______AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAU ((((((((((((.....))..)))))))((.((((((((...(((((.............)))))))))))))...))...........)))...... (-25.72 = -25.72 + 0.00)


| Location | 18,372,513 – 18,372,603 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18372513 90 + 22224390 GCCAGGCAAGCGA------AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAUUAAAAAGUUAACAUGUG--GACCGGGUUCGAGAAUUAGAG (((((((......------.)))))))...((((..(((((.......((..(((((((....)))))))..)).--.......)))))...)))).. ( -24.86) >DroSec_CAF1 17252 90 + 1 GACAGGCAAGCGA------AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAUUAAAAAGUUAACAUGUG--GACCGGGUUCGAGAAUUAGAG ..(((((......------.))))).....((((..(((((.......((..(((((((....)))))))..)).--.......)))))...)))).. ( -18.26) >DroYak_CAF1 16079 98 + 1 GCCAGGCAAGCGAAGCCGAAGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAUUAAAAAGUUAACAUGUGGGGACCGGGUCCGAGAAUUGGUG (((((((...((....))..)))))))..((..(..((......))...((((((((((....)))))))......((((...)))))))..)..)). ( -28.60) >consensus GCCAGGCAAGCGA______AGCCUGGCAAAUUGAAAUCGAAAAAGAAAACUCGUUAAUUAAAAAGUUAACAUGUG__GACCGGGUUCGAGAAUUAGAG (((((((.............)))))))...(((((..((.....((....))(((((((....)))))))..........))..)))))......... (-19.32 = -19.99 + 0.67)


| Location | 18,372,513 – 18,372,603 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18372513 90 - 22224390 CUCUAAUUCUCGAACCCGGUC--CACAUGUUAACUUUUUAAUUAACGAGUUUUCUUUUUCGAUUUCAAUUUGCCAGGCU------UCGCUUGCCUGGC .........(((((...((..--.((.((((((........)))))).))...))..))))).........(((((((.------......))))))) ( -20.70) >DroSec_CAF1 17252 90 - 1 CUCUAAUUCUCGAACCCGGUC--CACAUGUUAACUUUUUAAUUAACGAGUUUUCUUUUUCGAUUUCAAUUUGCCAGGCU------UCGCUUGCCUGUC .........(((((...((..--.((.((((((........)))))).))...))..)))))...........(((((.------......))))).. ( -14.90) >DroYak_CAF1 16079 98 - 1 CACCAAUUCUCGGACCCGGUCCCCACAUGUUAACUUUUUAAUUAACGAGUUUUCUUUUUCGAUUUCAAUUUGCCAGGCUUCGGCUUCGCUUGCCUGGC ....((..(((((((...))))......(((((........))))))))..))..................(((((((...(((...))).))))))) ( -22.20) >consensus CUCUAAUUCUCGAACCCGGUC__CACAUGUUAACUUUUUAAUUAACGAGUUUUCUUUUUCGAUUUCAAUUUGCCAGGCU______UCGCUUGCCUGGC .........(((((...((.....((.((((((........)))))).))...))..))))).........(((((((.............))))))) (-16.01 = -16.12 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:25 2006