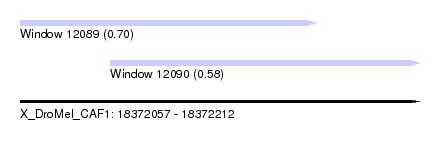
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,372,057 – 18,372,212 |
| Length | 155 |
| Max. P | 0.703054 |

| Location | 18,372,057 – 18,372,172 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
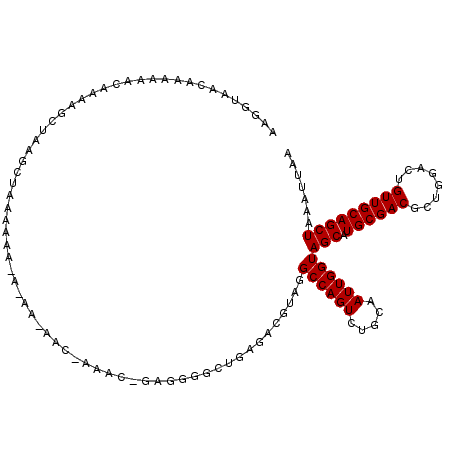
>X_DroMel_CAF1 18372057 115 + 22224390 AAGGUAACAAAAAACAAAAGCUAAGCUAAAAAA-ACAACAACAAAACAGAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAA ..................((((..((((.....-.......(((..((((..((((........))))..))))...))).))))..((((((......))))))))))....... ( -24.92) >DroSec_CAF1 16770 116 + 1 AAGGUAACAAAAAACAAAAGCUAAGCUAAAAAAAAAAAGAACUAAACUGAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAA ..................((((..((((..((.....((..((.......))..)).....(((((....)))))..))..))))..((((((......))))))))))....... ( -21.50) >DroEre_CAF1 15961 88 + 1 AAGGUAACAAAAAGCAAAAACGGAUGAAAAAA-----------------AA-----------CAAGCCAGUCUGCAAUUGGUAGCAUGCGACUCUGGACUGUUGCAGCUAAAUUAA ................................-----------------..-----------...((((((.....))))))(((.((((((........)))))))))....... ( -16.00) >consensus AAGGUAACAAAAAACAAAAGCUAAGCUAAAAAA_A_AA_AAC_AAAC_GAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAA .................................................................((((((.....))))))(((.((((((........)))))))))....... (-15.47 = -15.47 + 0.00)


| Location | 18,372,092 – 18,372,212 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -25.32 |
| Energy contribution | -27.43 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
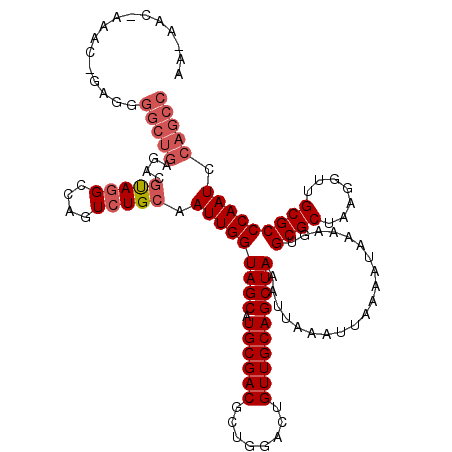
>X_DroMel_CAF1 18372092 120 + 22224390 AACAACAAAACAGAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAAAUUAAAAUAAAAGUGCGCUAAGGUUGCGCCCAAUCCAGCC ................(((((.((.(((((....)))))..((((((((.(((((((......)))))))))))....................((((.......))))))))))))))) ( -34.80) >DroSec_CAF1 16806 120 + 1 AAGAACUAAACUGAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAAAUUAAAAUAAAAGUGCGCUAAGGUUGCGCCCAAUGCAGCC ................(((((....(((((....))))).(((((((((.(((((((......)))))))))))....................((((.......))))))))).))))) ( -35.10) >DroEre_CAF1 15993 96 + 1 -------------AA-----------CAAGCCAGUCUGCAAUUGGUAGCAUGCGACUCUGGACUGUUGCAGCUAAAUUAAAUUAAAAUAAAAGUGCGCUAAGGUUGCGCCCAAUCCAGCC -------------..-----------.......(.(((..(((((((((.((((((........))))))))))....................((((.......))))))))).)))). ( -24.00) >consensus AA_AAC_AAAC_GAGGGGCUGAGACGUAGGCCAGUCUGCAAUUGGUAGCAUGCGACGCUGGACUGUUGCAGCUAAAUUAAAUUAAAAUAAAAGUGCGCUAAGGUUGCGCCCAAUCCAGCC ................(((((....(((((....))))).(((((((((.((((((........))))))))))....................((((.......))))))))).))))) (-25.32 = -27.43 + 2.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:23 2006