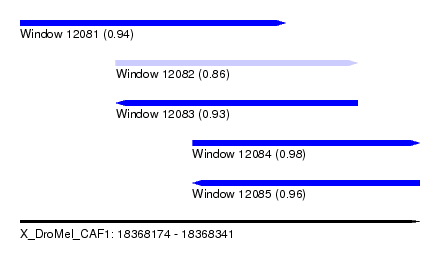
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,368,174 – 18,368,341 |
| Length | 167 |
| Max. P | 0.982508 |

| Location | 18,368,174 – 18,368,285 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.24 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -29.03 |
| Energy contribution | -28.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18368174 111 + 22224390 GCUGGCUGGUGGGUGAUAUAAUGUACAAAUCGCCAGAAUGGGGCC--------GAAAUGGCUGAAUGUCACAAGAAGGAAGAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUA .(((((.(((..(((........)))..))))))))..((((((.--------....(.(((....(((.((((..(((..((....)).)))..)))).)))))).).)))))).... ( -36.50) >DroSec_CAF1 12770 111 + 1 GCGGUCUGGUGGGUGAUACAAUGUACAAAUCGCCAGAAUGGAGCC--------GAAAUGGCUGAAUGUCACAAGAAGGAAGAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUA ..((((((((((....(((...)))....)))))))).((((((.--------....(.(((....(((.((((..(((..((....)).)))..)))).)))))).).)))))))).. ( -38.10) >DroYak_CAF1 11850 110 + 1 GCUGGCUGGUGGGUGAUA-------CAAAUCGGCAGAAUGGGGCCGAAAUGGCCAAAUGGCCAAAUGUCACCA--AGGAAAAGGAAACUAUCCGACUUGCGGCAGCGAAGCUCCACCUA (((.((((.(((((((((-------....(((((........)))))..(((((....)))))..))))))).--.(((..((....)).)))......)).))))..)))........ ( -44.20) >consensus GCUGGCUGGUGGGUGAUA_AAUGUACAAAUCGCCAGAAUGGGGCC________GAAAUGGCUGAAUGUCACAAGAAGGAAGAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUA .......(((((((((((...........((....))....((((.............))))...)))))).....(((..((....)).)))(((((((....)).)))))))))).. (-29.03 = -28.92 + -0.11)


| Location | 18,368,214 – 18,368,315 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -29.45 |
| Energy contribution | -30.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18368214 101 + 22224390 GGGCC--------GAAAUGGCUGAAUGUCACAAGAAGGAA-GAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACAAAGGGGCACAGGUGCGCCUACGCAGGUG .((((--------.....))))...((((.((((..(((.-.((....)).)))..)))).)))).........(((((.(..(((.(((....))).)))..).))))) ( -37.10) >DroSec_CAF1 12810 100 + 1 GAGCC--------GAAAUGGCUGAAUGUCACAAGAAGGAA-GAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUG .((((--------.....))))...((((.((((..(((.-.((....)).)))..)))).))))(((..(((((.......-.)))))..(((....))).)))..... ( -37.40) >DroEre_CAF1 12027 95 + 1 ------------CGAAAUGGACGAAUGUCACAA--AGGAAAAAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUG ------------.....(((((....))).)).--.(((...((....)).))).((((((((((.(...(((((.......-.)))))...).))).....))))))). ( -29.20) >DroYak_CAF1 11883 106 + 1 GGGCCGAAAUGGCCAAAUGGCCAAAUGUCACCA--AGGAA-AAGGAAACUAUCCGACUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUG ((.......(((((....))))).......)).--.(((.-.((....)).))).((((((((((.(...(((((.......-.)))))...).))).....))))))). ( -37.64) >consensus GGGCC________GAAAUGGCCGAAUGUCACAA__AGGAA_AAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA_AGGGGCACAGGUGCGCCUACGCAGGUG ..................((((...((((.((((..(((...((....)).)))..)))).))))....)))).(((((....(((.(((....))).)))....))))) (-29.45 = -30.95 + 1.50)


| Location | 18,368,214 – 18,368,315 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -29.14 |
| Energy contribution | -28.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18368214 101 - 22224390 CACCUGCGUAGGCGCACCUGUGCCCCUUUGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC-UUCCUUCUUGUGACAUUCAGCCAUUUC--------GGCCC ((((((((.(((.(((....))).))).))))))))..((......))((((((..(((.((......)-))))..)))))).......(((.....--------))).. ( -35.30) >DroSec_CAF1 12810 100 - 1 CACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC-UUCCUUCUUGUGACAUUCAGCCAUUUC--------GGCUC ((((((((..(((((....)))))...-))))))))(((((..((((.((((((..(((.((......)-))))..)))))).....))))......--------))))) ( -37.80) >DroEre_CAF1 12027 95 - 1 CACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUUUUUCCU--UUGUGACAUUCGUCCAUUUCG------------ ((((((((..(((((....)))))...-))))))))(((((...(((.(....).)))..)))))..........--.((.(((....))))).....------------ ( -30.80) >DroYak_CAF1 11883 106 - 1 CACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGUCGGAUAGUUUCCUU-UUCCU--UGGUGACAUUUGGCCAUUUGGCCAUUUCGGCCC ((((((((..(((((....)))))...-))))))))(.((......)))....((((((..(((.((..-.....--.)).)))...(((((....))))).)))))).. ( -40.10) >consensus CACCUGCGUAGGCGCACCUGUGCCCCU_UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC_UUCCU__UUGUGACAUUCAGCCAUUUC________GGCCC ((((((((.(((.(((....))).))).))))))))(((..((((.(.(....).)(((.....)))............))))..)))...................... (-29.14 = -28.95 + -0.19)


| Location | 18,368,246 – 18,368,341 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -38.71 |
| Energy contribution | -38.78 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18368246 95 + 22224390 -GAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACAAAGGGGCACAGGUGCGCCUACGCAGGUGGGCCGCAUGCUAAUAACAGUGCCUUA -((((..(((....(((.(((((((((...)))((((((.(..(((.(((....))).)))..).)))))))))))).)))......))).)))). ( -39.00) >DroSec_CAF1 12842 94 + 1 -GAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUGGGCCGCAUGCUAAUAACAGCGCCUUA -.((....))....(((.(((((((((...)))((((((.(.-(((.(((....))).)))..).)))))))))))).(((......))))))... ( -40.20) >DroEre_CAF1 12053 95 + 1 AAAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUGGGCCGCAUGCUAAUAACAGCGCCUUA ..((....))....(((.(((((((((...)))((((((.(.-(((.(((....))).)))..).)))))))))))).(((......))))))... ( -40.10) >DroYak_CAF1 11921 94 + 1 -AAGGAAACUAUCCGACUUGCGGCAGCGAAGCUCCACCUACA-AGGGGCACAGGUGCGCCUACGCAGGUGGGCCGCAUGCUAAUAACAGCGCCUUA -((((.............(((((((((...)))((((((.(.-(((.(((....))).)))..).)))))))))))).(((......))).)))). ( -35.80) >consensus _AAGGAAACUAUCCGGCUUGCGGCAGCGAAGCUCCACCUACA_AGGGGCACAGGUGCGCCUACGCAGGUGGGCCGCAUGCUAAUAACAGCGCCUUA ..((....))....(((.(((((((((...)))((((((....(((.(((....))).)))....)))))))))))).(((......))))))... (-38.71 = -38.78 + 0.06)

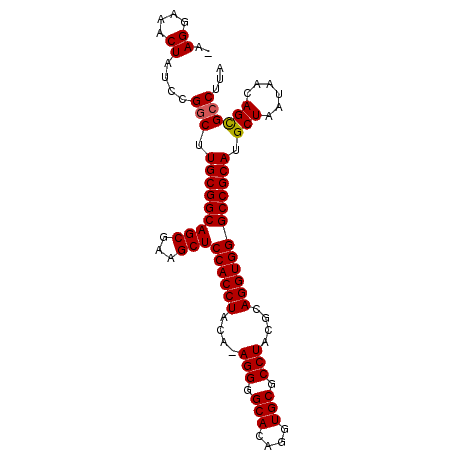
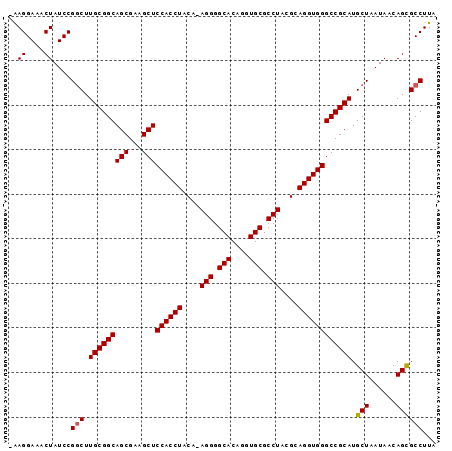
| Location | 18,368,246 – 18,368,341 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -46.75 |
| Consensus MFE | -46.09 |
| Energy contribution | -46.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18368246 95 - 22224390 UAAGGCACUGUUAUUAGCAUGCGGCCCACCUGCGUAGGCGCACCUGUGCCCCUUUGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC- ..(((.((((((....((.(((((((((((((((.(((.(((....))).))).)))))))))(((...))))))))).))..))))))..))).- ( -45.10) >DroSec_CAF1 12842 94 - 1 UAAGGCGCUGUUAUUAGCAUGCGGCCCACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC- ...(((((((....)))).(((((((((((((((..(((((....)))))...-)))))))))(((...))))))))).)))(((.....)))..- ( -48.20) >DroEre_CAF1 12053 95 - 1 UAAGGCGCUGUUAUUAGCAUGCGGCCCACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUUU ...(((((((....)))).(((((((((((((((..(((((....)))))...-)))))))))(((...))))))))).)))(((.....)))... ( -48.20) >DroYak_CAF1 11921 94 - 1 UAAGGCGCUGUUAUUAGCAUGCGGCCCACCUGCGUAGGCGCACCUGUGCCCCU-UGUAGGUGGAGCUUCGCUGCCGCAAGUCGGAUAGUUUCCUU- ...(((((((....)))).(((((((((((((((..(((((....)))))...-)))))))))(((...))))))))).)))(((.....)))..- ( -45.50) >consensus UAAGGCGCUGUUAUUAGCAUGCGGCCCACCUGCGUAGGCGCACCUGUGCCCCU_UGUAGGUGGAGCUUCGCUGCCGCAAGCCGGAUAGUUUCCUC_ ...(((((((....)))).(((((((((((((((.(((.(((....))).))).)))))))))(((...))))))))).)))(((.....)))... (-46.09 = -46.15 + 0.06)


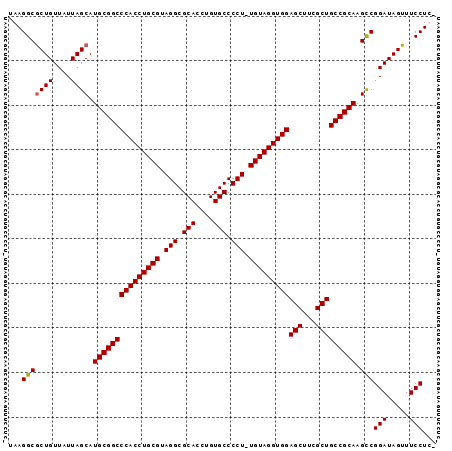
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:18 2006