
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,365,652 – 18,365,769 |
| Length | 117 |
| Max. P | 0.965449 |

| Location | 18,365,652 – 18,365,742 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -20.72 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18365652 90 + 22224390 AAAAAAGGACCGAUGAUAGUCCUGCGGCUGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....((((((((((((((......)))))))..)))))))(((.........)))...)))((((.(....))))).. ( -23.50) >DroSec_CAF1 10287 90 + 1 AAAAAAGGACCGAUGAUAGUCCUGCGGCUGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....((((((((((((((......)))))))..)))))))(((.........)))...)))((((.(....))))).. ( -23.50) >DroSim_CAF1 11147 90 + 1 AAAAAAGGACCGAUGAUAGUCCUGCGGCUGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....((((((((((((((......)))))))..)))))))(((.........)))...)))((((.(....))))).. ( -23.50) >DroEre_CAF1 9437 90 + 1 AAAAAAGGACCGAUGAUAGUCCUGCGGCAGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....(((((((.((((((.........)))))))))))))(((.........)))...)))((((.(....))))).. ( -23.20) >DroYak_CAF1 9422 90 + 1 UAAAAAGGACCGAUGAUAGUCCUGCGGCAGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....(((((((.((((((.........)))))))))))))(((.........)))...)))((((.(....))))).. ( -23.20) >consensus AAAAAAGGACCGAUGAUAGUCCUGCGGCUGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAU .........(((....((((((((((((((......)))))))..)))))))(((.........)))...)))................. (-20.72 = -21.12 + 0.40)

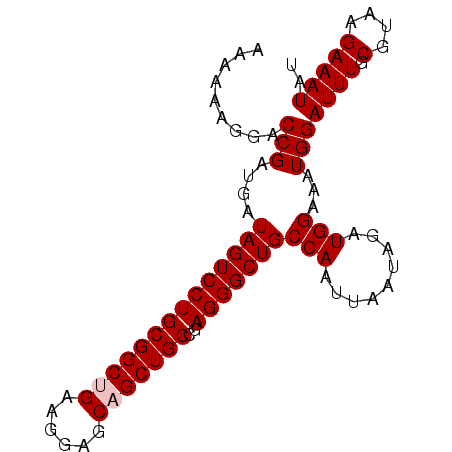
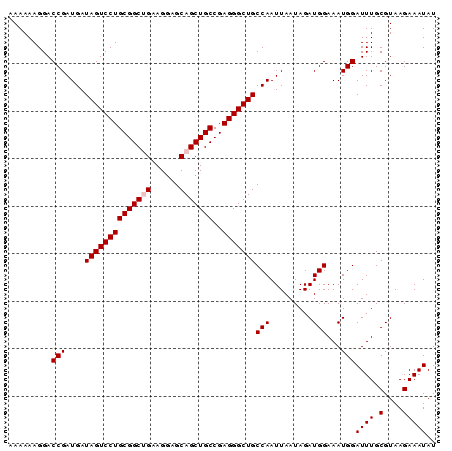
| Location | 18,365,652 – 18,365,742 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18365652 90 - 22224390 AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCAGCCGCAGGACUAUCAUCGGUCCUUUUUU ...........((..........(((.........)))..(((...))).((((......)))).))((((((......))))))..... ( -16.50) >DroSec_CAF1 10287 90 - 1 AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCAGCCGCAGGACUAUCAUCGGUCCUUUUUU ...........((..........(((.........)))..(((...))).((((......)))).))((((((......))))))..... ( -16.50) >DroSim_CAF1 11147 90 - 1 AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCAGCCGCAGGACUAUCAUCGGUCCUUUUUU ...........((..........(((.........)))..(((...))).((((......)))).))((((((......))))))..... ( -16.50) >DroEre_CAF1 9437 90 - 1 AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCUGCCGCAGGACUAUCAUCGGUCCUUUUUU ....................................(((((.....(((....)))....)))))..((((((......))))))..... ( -17.60) >DroYak_CAF1 9422 90 - 1 AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCUGCCGCAGGACUAUCAUCGGUCCUUUUUA ....................................(((((.....(((....)))....)))))..((((((......))))))..... ( -17.60) >consensus AUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCAGCCGCAGGACUAUCAUCGGUCCUUUUUU ...........((...........((.........))(((((........)))))..........))((((((......))))))..... (-15.70 = -15.70 + 0.00)


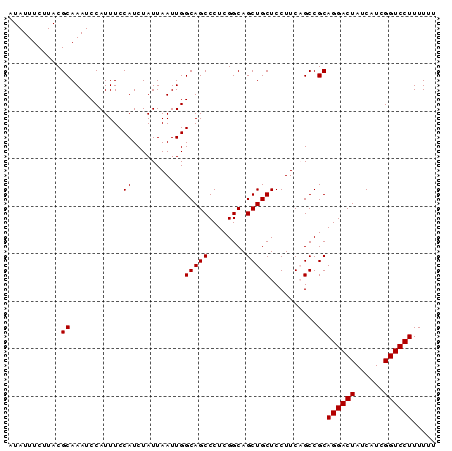
| Location | 18,365,677 – 18,365,769 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18365677 92 + 22224390 GGC---------------UGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGUUUCCACCCCCCCCCCCC------------- ((.---------------.(..((.((((((......))))))...........(((((((((...((((((.....))))))...)))))))))))..)..))...------------- ( -29.90) >DroSec_CAF1 10312 84 + 1 GGC---------------UGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGCUUCCACCC--------C------------- ...---------------....((.((((((......))))))...........(((((.(((...((((((.....))))))...))).))))))).--------.------------- ( -25.00) >DroAna_CAF1 13036 99 + 1 GGCAGAGGCUUCACUUUCUGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCAUUUCCACUUU--------------------- (((((..(((((..........)))))..)))))..((....))..........(((((((((...((((((.....))))))...)))))))))....--------------------- ( -36.70) >DroSim_CAF1 11172 85 + 1 GGC---------------UGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGCUUCCACCCC-------C------------- ...---------------....((.((((((......))))))...........(((((.(((...((((((.....))))))...))).)))))))..-------.------------- ( -25.00) >DroEre_CAF1 9462 85 + 1 GGC---------------AGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGCUACCACUUCC-------------------- ...---------------.(((((.((((((......)))))))..........(((...(((...((((((.....))))))...)))...))))))).-------------------- ( -23.20) >DroYak_CAF1 9447 105 + 1 GGC---------------AGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGCUUCCACCCCCCUUUCCUGCCCAUACCCAUA (((---------------((((((.((((((......))))))...........(((((.(((...((((((.....))))))...))).)))))....))))..))))).......... ( -35.20) >consensus GGC_______________UGAAGGAGCAGCUGCCGAGGGCUGCCAAUUAAUAGAUGGAAAUGGAUUUGCGUAAGAAAUAUGCGUUGCCGCUUCCACCCC_______C_____________ ......................((.((((((......))))))...........(((((.(((...((((((.....))))))...))).)))))))....................... (-23.99 = -23.80 + -0.19)

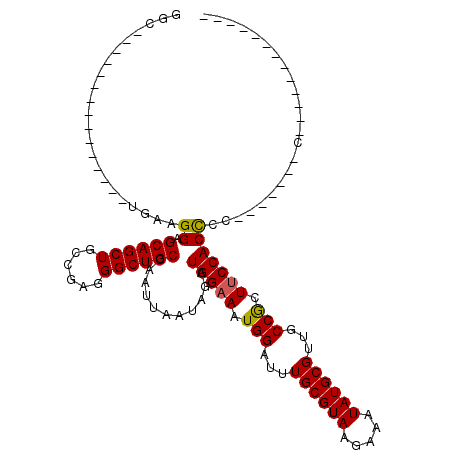
| Location | 18,365,677 – 18,365,769 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.41 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18365677 92 - 22224390 -------------GGGGGGGGGGGGUGGAAACGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCA---------------GCC -------------(((((((((..(((((...(....)((.((.....)).))...)))))..))...........(((.(((...))).))).))))))).---------------... ( -31.50) >DroSec_CAF1 10312 84 - 1 -------------G--------GGGUGGAAGCGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCA---------------GCC -------------.--------(((((((((((....)))...........(......)..)))))))).......((((((........))))))......---------------... ( -22.80) >DroAna_CAF1 13036 99 - 1 ---------------------AAAGUGGAAAUGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCAGAAAGUGAAGCCUCUGCC ---------------------...((((((((((....((.((.....)).))....)))))))))).........(((((.....(((.(((..((.....)).)))...))).))))) ( -25.80) >DroSim_CAF1 11172 85 - 1 -------------G-------GGGGUGGAAGCGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCA---------------GCC -------------(-------(..(((((.(((....))).((.....))......)))))..))...........((((((........))))))......---------------... ( -26.10) >DroEre_CAF1 9462 85 - 1 --------------------GGAAGUGGUAGCGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCU---------------GCC --------------------((((((((..(((....))).((.....)).......))))))))...........(((((.....(((....)))....))---------------))) ( -24.90) >DroYak_CAF1 9447 105 - 1 UAUGGGUAUGGGCAGGAAAGGGGGGUGGAAGCGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCU---------------GCC ..........(((((((...((..(((((.(((....))).((.....))......)))))..))...........((((((........))))))..))))---------------))) ( -35.60) >consensus _____________G_______GGGGUGGAAGCGGCAACGCAUAUUUCUUACGCAAAUCCAUUUCCAUCUAUUAAUUGGCAGCCCUCGGCAGCUGCUCCUUCA_______________GCC ......................(((((((((.((....((.((.....)).))....)).))))))))).......((((((........))))))........................ (-20.41 = -20.47 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:09 2006