
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,359,574 – 18,359,691 |
| Length | 117 |
| Max. P | 0.998230 |

| Location | 18,359,574 – 18,359,665 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 68.46 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
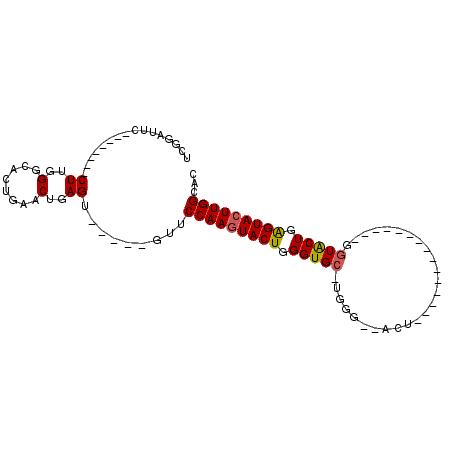
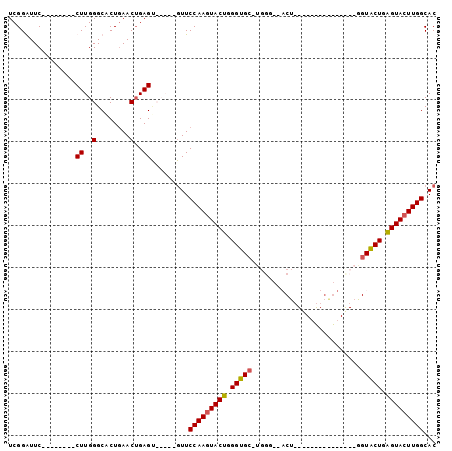
>X_DroMel_CAF1 18359574 91 + 22224390 UCGGAUUC--------CUUGGGCACUGAACAGAGUUCGCAGUUCCAAAUACUGGGUGC-UGGGUAACU-GGU-CUGAGUACUGAGUACUGAGUACUUGGCAC .(((((((--------..((..(...)..)))))))))..((.((((.((((.(((((-(.((((.((-...-...)))))).)))))).)))).)))).)) ( -28.70) >DroSim_CAF1 4699 71 + 1 UCGGAUUC--------CUUGGGCACUGAACUGAGCUCGGAGUUCCAAGUACUGGGUGC-UGGG----------------------UACUGAGUACUUGGCAC ..((((((--------(..((((.(......).)))))))))))((((((((.(((((-...)----------------------)))).)))))))).... ( -29.00) >DroEre_CAF1 4418 71 + 1 UCGGGUUC--------CUUGGGCACUCAACUGAG-------UUCCAAGUACUGGGUGCCUGGU-GCCU---------------GGUGCUGGGUACUUGGCAC .(.(((..--------((((((.((((....)))-------))))))).))).)(((((.(((-((((---------------......))))))).))))) ( -32.90) >DroYak_CAF1 4762 95 + 1 UCGGGUUCCUUGCUUCCUUGGGCGCUGAACUGAGU-----GUUCCAAGUACUUGGUGC-UGGU-UACUGGGUAUUGGGUAUUGGGUGCUGGGUACUUGGCAC ...(((((..((((......))))..)))))..((-----(..(((((((((..(..(-(.(.-((((.(....).)))).).))..)..)))))))))))) ( -36.00) >consensus UCGGAUUC________CUUGGGCACUGAACUGAGU_____GUUCCAAGUACUGGGUGC_UGGG__ACU_______________GGUACUGAGUACUUGGCAC ................((..(........)..)).........(((((((((.(((((..........................))))).)))))))))... (-17.37 = -17.37 + -0.00)

| Location | 18,359,595 – 18,359,691 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.82 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
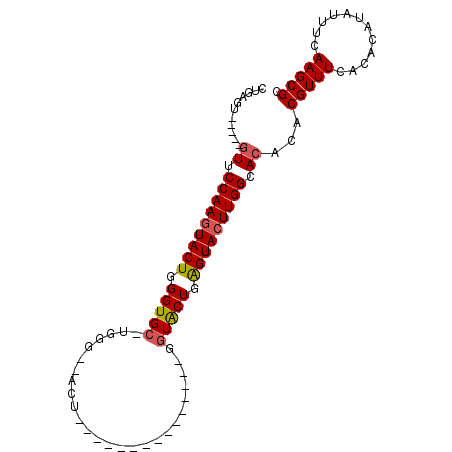
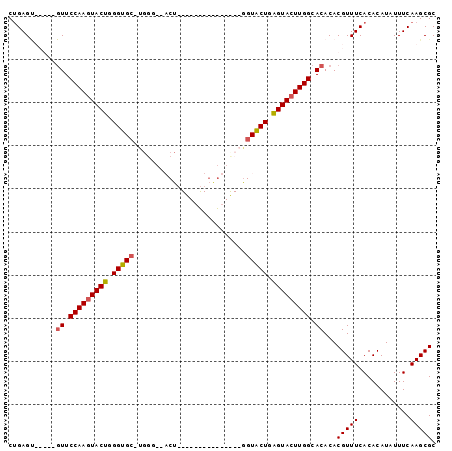
>X_DroMel_CAF1 18359595 96 + 22224390 CAGAGUUCGCAGUUCCAAAUACUGGGUGC-UGGGUAACU-GGU-CUGAGUACUGAGUACUGAGUACUUGGCACACACGUUUCACACAUAUUUCAAGCGC .......(((.((.((((.((((.(((((-(.((((.((-...-...)))))).)))))).)))).)))).))....((.....)).........))). ( -27.60) >DroSim_CAF1 4720 76 + 1 CUGAGCUCGGAGUUCCAAGUACUGGGUGC-UGGG----------------------UACUGAGUACUUGGCACACACGUUUCACACAUAUUUCAAGCGC ....(((.(((((.(((((((((.(((((-...)----------------------)))).))))))))).))....((.....))....))).))).. ( -25.40) >DroEre_CAF1 4439 76 + 1 CUGAG-------UUCCAAGUACUGGGUGCCUGGU-GCCU---------------GGUGCUGGGUACUUGGCACACACGUUUCACACAUAUUUCAAGCGC ....(-------(.(((((((((.((..((.(..-..).---------------))..)).))))))))).))...(((((............))))). ( -27.50) >DroYak_CAF1 4791 92 + 1 CUGAGU-----GUUCCAAGUACUUGGUGC-UGGU-UACUGGGUAUUGGGUAUUGGGUGCUGGGUACUUGGCACACACGUUUCACACAUAUUUCAAGCGC ....((-----((.(((((((((..(..(-(.(.-((((.(....).)))).).))..)..))))))))).)))).(((((............))))). ( -33.80) >consensus CUGAGU_____GUUCCAAGUACUGGGUGC_UGGG__ACU_______________GGUACUGAGUACUUGGCACACACGUUUCACACAUAUUUCAAGCGC ...........((.(((((((((.(((((..........................))))).))))))))).))...(((((............))))). (-17.12 = -17.37 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:37:00 2006