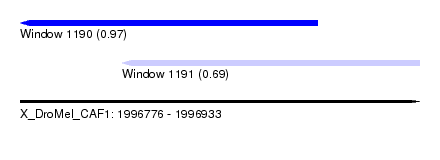
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,996,776 – 1,996,933 |
| Length | 157 |
| Max. P | 0.972865 |

| Location | 1,996,776 – 1,996,893 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1996776 117 - 22224390 UCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUUUUGCACCCCAUAAA-AAACCUUUUCCGGUGGUGCGCCCAUUU--UUUAUUCGCUUCGAUCUUCCGUCGGCAUUUUGUACUGCCAA ...........(((((.((((.(((((..........(((((((..(((-(....))))..)).)))))........--...........((((.....))))))))))))).))))).. ( -30.10) >DroSec_CAF1 28793 118 - 1 UCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU--UUUAUUCGCUUCGAUCUUCCGUCGGCAUUUUAAACUGCCAA .......((.(((((((((....))))..........(((((((..((((.....))))..)).)))))........--......)))))))..........((((........)))).. ( -28.90) >DroSim_CAF1 23002 118 - 1 UCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU--UUUAUUCGCUUCGAUCUUCCGUCGGCAUUUUAAACUGCCAA .......((.(((((((((....))))..........(((((((..((((.....))))..)).)))))........--......)))))))..........((((........)))).. ( -28.90) >DroEre_CAF1 18573 119 - 1 UCUCCCUCGCAGGCGGCACCAUGGUGCUAUCUUUGUUGCACCCCAUAAA-AAACCUCUUUCGGUGGUGUGCCCAUUUUUUUUAUUCGUUUCGAUCUUCCGUCGGCAUUUUGUGCUGCCAA ...........((((((((...(((((..........((((.((((..(-((.....)))..)))).))))...................((((.....)))))))))..)))))))).. ( -33.80) >DroYak_CAF1 29323 117 - 1 UCUCUCCC-CAGGCGGCACAAUGGUGCUGUCUCUGUUGCACUCCAUAAAAAAACCUCUUUCGGUGAUGCGCCCAUUU--UUUAUUCGUUUCGAUCUUCCGUCGGCAUUUUGUGCUGCCAA ........-..((((((((((.(((((.(((.(((.........................))).)))))))).....--..........(((((.....)))))....)))))))))).. ( -31.51) >consensus UCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU__UUUAUUCGCUUCGAUCUUCCGUCGGCAUUUUGUACUGCCAA ...........((((((((....))))).........(((((((.................)).))))))))...................(((.....)))((((........)))).. (-25.39 = -25.27 + -0.12)

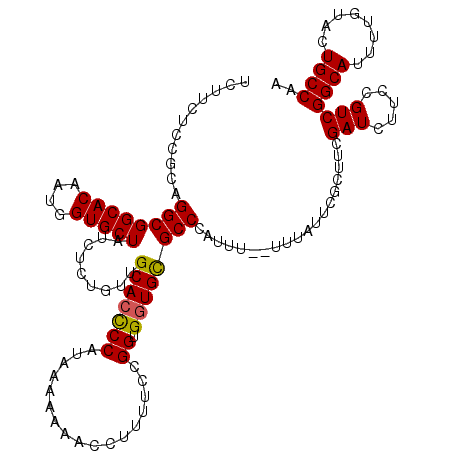
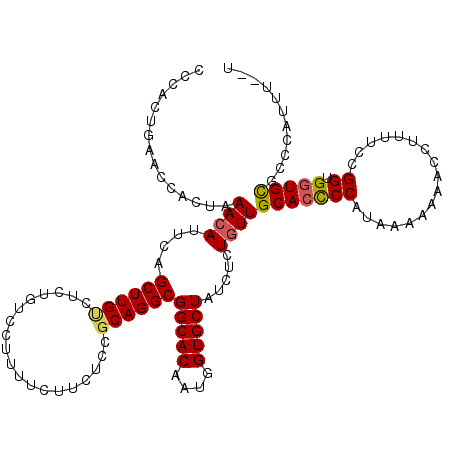
| Location | 1,996,816 – 1,996,933 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1996816 117 - 22224390 CCCACUGAACCACUAAACAUUUAGCUUGUCUCUGUCCUAUUCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUUUUGCACCCCAUAAA-AAACCUUUUCCGGUGGUGCGCCCAUUU--U ..((..((....((((....))))....))..)).................((((((((....))))).........(((((((..(((-(....))))..)).)))))))).....--. ( -23.80) >DroSec_CAF1 28833 118 - 1 CCCACUGAACCACUAAACAUUCAGCUUGCCUCUGUCCUUUUCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU--U ...............((((....((((((...................))))))(((((....))))).....))))(((((((..((((.....))))..)).)))))........--. ( -25.61) >DroSim_CAF1 23042 118 - 1 CCCACUGAACCACUAAACAUUCAGCUUGCCUUUGUCCUUUUCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU--U ...............((((....((((((...................))))))(((((....))))).....))))(((((((..((((.....))))..)).)))))........--. ( -25.61) >DroEre_CAF1 18613 119 - 1 CCCACUGAACCACUAAACAGCCAGCUUGUCUCUAUCCCUUUCUCCCUCGCAGGCGGCACCAUGGUGCUAUCUUUGUUGCACCCCAUAAA-AAACCUCUUUCGGUGGUGUGCCCAUUUUUU .((((((((..........(((.((((((...................))))))))).....(((((..........))))).......-........)))))))).............. ( -29.01) >DroYak_CAF1 29363 117 - 1 CCCACACAACCACUAAACACCCAGCUUGUCUCUGUCCCUUUCUCUCCC-CAGGCGGCACAAUGGUGCUGUCUCUGUUGCACUCCAUAAAAAAACCUCUUUCGGUGAUGCGCCCAUUU--U .................(((((((.......)))..............-.(((((((((....))))))))).............................))))............--. ( -18.40) >consensus CCCACUGAACCACUAAACAUUCAGCUUGUCUCUGUCCUUUUCUUCUCCGCAGGCGGCACAAUGGUGCUAUCUCUGUUGCACCCCAUAAAAAAACCUUUUCCGGUGGUGCGCCCAUUU__U ...............((((....((((((...................))))))(((((....))))).....))))(((((((.................)).)))))........... (-20.46 = -20.58 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:31 2006