
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,338,650 – 18,338,804 |
| Length | 154 |
| Max. P | 0.996027 |

| Location | 18,338,650 – 18,338,770 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -37.34 |
| Energy contribution | -37.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
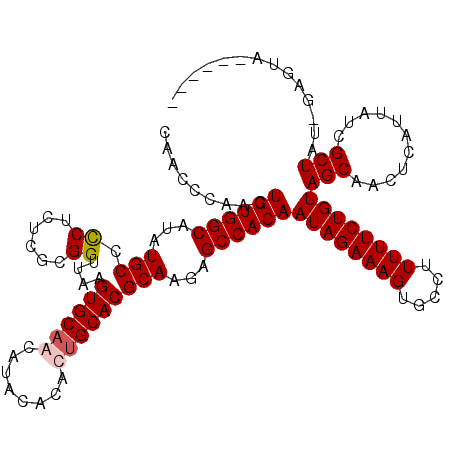
>X_DroMel_CAF1 18338650 120 + 22224390 GGGACUUGAGGCCGAGAUAUAUCCAUGUCGGCACUCCUUGCAACCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACACAGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).)))(((((.....(((((((((....)))......))))))..((((............)))))))))............)))).)) ( -37.40) >DroSec_CAF1 16596 120 + 1 GGGACUUGAGGCCGAGGUAUAUCCAUGUCGGCACUCCUUGCAACCCAACUGUGGCAUAUGCCUCUCUCGCGGUUAAGUGCAACAUACACACUGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).)))((((((((((....(.(((....))).)....).))))..(((((..........))))))))))............)))).)) ( -39.40) >DroSim_CAF1 16423 120 + 1 GGGACUUGAGGCCGAGGUAUAUCCAUGUCGGCACUCCUUGCAACCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACACUGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).)))(((((.....(((((((((....)))......))))))..(((((..........))))))))))............)))).)) ( -39.00) >DroEre_CAF1 16219 120 + 1 GGGACUUGAGGCCGAGGUAUAUCCAUGUCGGCACUCCUUGCAAUCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACAUUGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).)))(((((.....(((((((((....)))......))))))..((((((........)))))))))))............)))).)) ( -40.40) >DroYak_CAF1 16632 120 + 1 GGGACUUGAGGCCGAGGUAUAUCCAUGUCGGCACUCCUUGCAAUCCAACUGUGGCAUAUGCACCUCUCGCGGUUAAGUGCAACAUACACAUUGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).))).............((((((...((((((......)))...((((((........)))))))))...)))))).....)))).)) ( -40.60) >consensus GGGACUUGAGGCCGAGGUAUAUCCAUGUCGGCACUCCUUGCAACCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACACUGCACGCAAGAGCCACAAUAGAAAGUGCC ((.((((((((((((.((......)).))))).)))(((((.....(((((((((....)))......))))))..(((((..........))))))))))............)))).)) (-37.34 = -37.58 + 0.24)

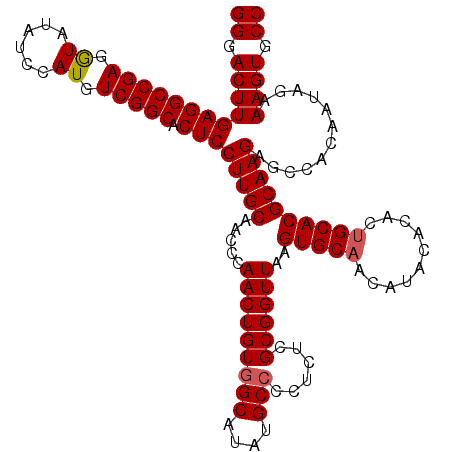
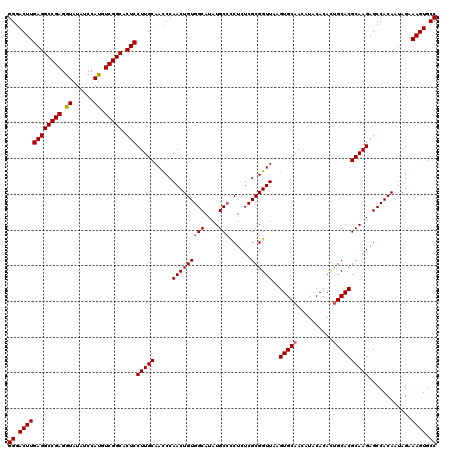
| Location | 18,338,650 – 18,338,770 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -44.46 |
| Consensus MFE | -41.20 |
| Energy contribution | -40.76 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18338650 120 - 22224390 GGCACUUUCUAUUGUGGCUCUUGCGUGCUGUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGGUUGCAAGGAGUGCCGACAUGGAUAUAUCUCGGCCUCAAGUCCC ((.((((....(((..((((......((((((((((((((.(.(((......))).).)))))))).)))))).))))..))).(((.(((((.(((....))).)))))))))))))). ( -42.90) >DroSec_CAF1 16596 120 - 1 GGCACUUUCUAUUGUGGCUCUUGCGUGCAGUGUGUAUGUUGCACUUAACCGCGAGAGAGGCAUAUGCCACAGUUGGGUUGCAAGGAGUGCCGACAUGGAUAUACCUCGGCCUCAAGUCCC .((((((...(((((..((((((((...((((((.....))))))....)))))))).(((....)))))))).))).)))...(((.(((((...((.....))))))))))....... ( -41.90) >DroSim_CAF1 16423 120 - 1 GGCACUUUCUAUUGUGGCUCUUGCGUGCAGUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGGUUGCAAGGAGUGCCGACAUGGAUAUACCUCGGCCUCAAGUCCC .((((((...((((((((...(((((((((........))))))....((.(....))))))...)))))))).))).)))...(((.(((((...((.....))))))))))....... ( -44.10) >DroEre_CAF1 16219 120 - 1 GGCACUUUCUAUUGUGGCUCUUGCGUGCAAUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGAUUGCAAGGAGUGCCGACAUGGAUAUACCUCGGCCUCAAGUCCC .(((..(((.((((((((...((((((((((......)))))))....((.(....))))))...)))))))).))).)))...(((.(((((...((.....))))))))))....... ( -46.90) >DroYak_CAF1 16632 120 - 1 GGCACUUUCUAUUGUGGCUCUUGCGUGCAAUGUGUAUGUUGCACUUAACCGCGAGAGGUGCAUAUGCCACAGUUGGAUUGCAAGGAGUGCCGACAUGGAUAUACCUCGGCCUCAAGUCCC .(((..(((.((((((((...((((((((((......)))))))...(((......))))))...)))))))).))).)))...(((.(((((...((.....))))))))))....... ( -46.50) >consensus GGCACUUUCUAUUGUGGCUCUUGCGUGCAGUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGGUUGCAAGGAGUGCCGACAUGGAUAUACCUCGGCCUCAAGUCCC .(((..(((.((((((((...((((((((((......))))))).....(....)....)))...)))))))).))).)))...(((.(((((...((.....))))))))))....... (-41.20 = -40.76 + -0.44)

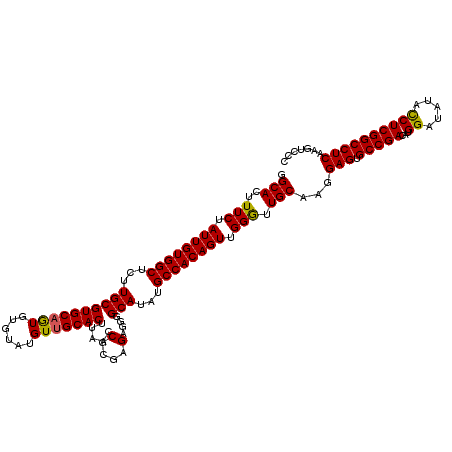
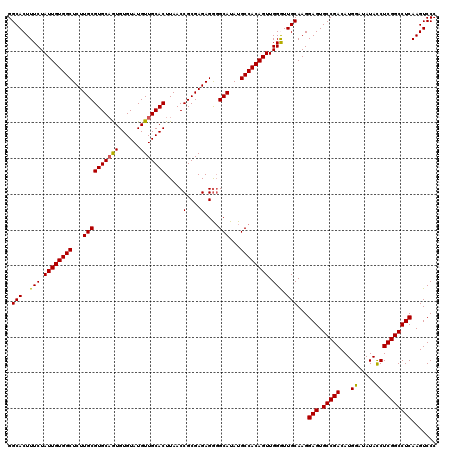
| Location | 18,338,690 – 18,338,804 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18338690 114 + 22224390 CAACCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACACAGCACGCAAGAGCCACAAUAGAAAGUGCCUUUUUCUGUAGCAACUCAUUAUCGCUUUGGAGUA------ ..((((((.((((((.........(((.((((((..(((.......)))..))).))).)))))))))((((((((.....)))))))).................)))).)).------ ( -26.90) >DroSec_CAF1 16636 113 + 1 CAACCCAACUGUGGCAUAUGCCUCUCUCGCGGUUAAGUGCAACAUACACACUGCACGCAAGAGCCACAAUAGAAAGUGCCUUUUUCUGUAGCAACUCAUUAUCGCUAU-GAGUA------ .........((((((.............(((((...(((.......)))))))).(....).))))))((((((((.....))))))))....((((((.......))-)))).------ ( -29.50) >DroEre_CAF1 16259 113 + 1 CAAUCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACAUUGCACGCAAGAGCCACAAUAGAAAGUGCCUUUUUCUGUAGCAAUUCAUUAUCGCUAU-GAGUA------ .........((((((...(((.((......))....((((((........)))))))))...))))))((((((((.....))))))))....((((((.......))-)))).------ ( -28.40) >DroYak_CAF1 16672 119 + 1 CAAUCCAACUGUGGCAUAUGCACCUCUCGCGGUUAAGUGCAACAUACACAUUGCACGCAAGAGCCACAAUAGAAAGUGCCUUUUUCUGUAGCAAUUCAUUGUUGCUAU-GAGUAUGAGUA .........((((((...((((((......)))...((((((........)))))))))...))))))..((((((.....))))))((((((((.....))))))))-........... ( -33.20) >consensus CAACCCAACUGUGGCAUAUGCCCCUCUCGCGGUUAAGUGCAACAUACACACUGCACGCAAGAGCCACAAUAGAAAGUGCCUUUUUCUGUAGCAACUCAUUAUCGCUAU_GAGUA______ .........((((((...(((.((......))....((((((........)))))))))...))))))((((((((.....))))))))(((...........))).............. (-24.19 = -24.75 + 0.56)

| Location | 18,338,690 – 18,338,804 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.38 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18338690 114 - 22224390 ------UACUCCAAAGCGAUAAUGAGUUGCUACAGAAAAAGGCACUUUCUAUUGUGGCUCUUGCGUGCUGUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGGUUG ------...(((((.((((((..((((.(((.........)))))))..))))))..((((((((.(.((.((((.....)))).)).))))))))).(((....)))....)))))... ( -34.50) >DroSec_CAF1 16636 113 - 1 ------UACUC-AUAGCGAUAAUGAGUUGCUACAGAAAAAGGCACUUUCUAUUGUGGCUCUUGCGUGCAGUGUGUAUGUUGCACUUAACCGCGAGAGAGGCAUAUGCCACAGUUGGGUUG ------.((((-(..((((((..((((.(((.........)))))))..))))))..((((((((...((((((.....))))))....)))))))).(((....))).....))))).. ( -34.70) >DroEre_CAF1 16259 113 - 1 ------UACUC-AUAGCGAUAAUGAAUUGCUACAGAAAAAGGCACUUUCUAUUGUGGCUCUUGCGUGCAAUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGAUUG ------.....-.(((((((.....)))))))............(..(((((((((((...((((((((((......)))))))....((.(....))))))...))))))).))))..) ( -36.40) >DroYak_CAF1 16672 119 - 1 UACUCAUACUC-AUAGCAACAAUGAAUUGCUACAGAAAAAGGCACUUUCUAUUGUGGCUCUUGCGUGCAAUGUGUAUGUUGCACUUAACCGCGAGAGGUGCAUAUGCCACAGUUGGAUUG ...........-.((((((.......))))))............(..(((((((((((...((((((((((......)))))))...(((......))))))...))))))).))))..) ( -34.50) >consensus ______UACUC_AUAGCGAUAAUGAAUUGCUACAGAAAAAGGCACUUUCUAUUGUGGCUCUUGCGUGCAAUGUGUAUGUUGCACUUAACCGCGAGAGGGGCAUAUGCCACAGUUGGAUUG .............(((((((.....)))))))............(..(((((((((((...((((((((((......))))))).....(....)....)))...))))))).))))..) (-30.44 = -30.38 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:44 2006