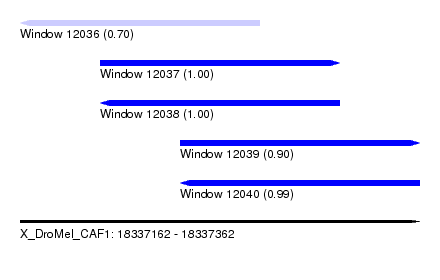
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,337,162 – 18,337,362 |
| Length | 200 |
| Max. P | 0.999979 |

| Location | 18,337,162 – 18,337,282 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -35.68 |
| Energy contribution | -35.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18337162 120 - 22224390 UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACAGUCAUCACAAUGAUACGACUCCAACUGCAGACGCAUAUUCGGAGACAAUCGAUC ((((....(((((((....)))))))...........(((((.(((.....))))))))))))...(((((....))))).(((((((...(((....))).....))))....)))... ( -35.70) >DroSec_CAF1 15113 120 - 1 UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGACACGACUCCAACUGCAGACGCAUACUCGGAGACAAUCGAUC ((((....(((((((....)))))))...........(((((.(((.....)))))))))))).(((((((....))))).)).((((...(((....))).....)))).......... ( -38.80) >DroSim_CAF1 14947 120 - 1 UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGACACGACUCCAACUGCAGACGCAUACUCGGAGACAAUCGAUC ((((....(((((((....)))))))...........(((((.(((.....)))))))))))).(((((((....))))).)).((((...(((....))).....)))).......... ( -38.80) >DroEre_CAF1 14746 120 - 1 UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGGGCAGCUGGCUCCGCGAACGGUCAUCACAAUGACACGGCUCCAACUGCAGACGCAUACUCGGAGACAAUCGAUC (((...(((((((((....)))))))...........(((((.((((((.....))))))....(((((((....))))).)))))))..........))......(....)...))).. ( -40.30) >DroYak_CAF1 15117 120 - 1 UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAGCAUGGAGGAGCAUGGAGCAGCUGGCUCCGCGAACGGUCAUCACAAUGACACGACGCCCACUGUAGACGCAUACUCGGAGACAAUCGAUC ..(((((((((((((....))))))).....((((......)))).....))))))((((((..(((((((....))))).)).))).....(((......)))..)))........... ( -37.90) >consensus UCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGACACGACUCCAACUGCAGACGCAUACUCGGAGACAAUCGAUC ((((....(((((((....)))))))...........(((((.(((.....)))))))))))).(((((((....))))).)).((((...(((....))).....)))).......... (-35.68 = -35.60 + -0.08)

| Location | 18,337,202 – 18,337,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -41.24 |
| Consensus MFE | -40.72 |
| Energy contribution | -40.40 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.22 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18337202 120 + 22224390 AUCAUUGUGAUGACUGUUCGCGGAGCCAGCGCCUCCGUGCUCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUC ....((((((((((.(((.(((((((((((.....((((......)))).....((((.((....)).))))))))))....))))).)))))))))))))................... ( -40.20) >DroSec_CAF1 15153 120 + 1 GUCAUUGUGAUGACCGUUCGCGGAGCCAGCGCCUCCGUGCUCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUC (.(.((((((((((.(((.(((((((((((.....((((......)))).....((((.((....)).))))))))))....))))).))))))))))))).).)............... ( -41.20) >DroSim_CAF1 14987 120 + 1 GUCAUUGUGAUGACCGUUCGCGGAGCCAGCGCCUCCGUGCUCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUC (.(.((((((((((.(((.(((((((((((.....((((......)))).....((((.((....)).))))))))))....))))).))))))))))))).).)............... ( -41.20) >DroEre_CAF1 14786 120 + 1 GUCAUUGUGAUGACCGUUCGCGGAGCCAGCUGCCCCGUGCUCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCAGACGUCAUCACAACGUCGUGCUUAAAAUAAUC (.(.((((((((((.((..(((((((((((.....((((......)))).....((((.((....)).))))))))))....)))))..)))))))))))).).)............... ( -42.70) >DroYak_CAF1 15157 120 + 1 GUCAUUGUGAUGACCGUUCGCGGAGCCAGCUGCUCCAUGCUCCUCCAUGCUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUC (.(.((((((((((.(((.(((((((((((.....((((......)))).....((((.((....)).))))))))))....))))).))))))))))))).).)............... ( -40.90) >consensus GUCAUUGUGAUGACCGUUCGCGGAGCCAGCGCCUCCGUGCUCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUC ....((((((((((.(((.(((((((((((.....((((......)))).....((((.((....)).))))))))))....))))).)))))))))))))................... (-40.72 = -40.40 + -0.32)


| Location | 18,337,202 – 18,337,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -46.98 |
| Consensus MFE | -45.66 |
| Energy contribution | -45.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.21 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18337202 120 - 22224390 GAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACAGUCAUCACAAUGAU .................((((((((((((...(((.......))).(((((((((....)))))))...........(((((.(((.....)))))))))).....)))))))))))).. ( -47.30) >DroSec_CAF1 15153 120 - 1 GAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGAC .................((((((((((((.(((((.......))).(((((((((....)))))))...........(((((.(((.....))))))))))))...)))))))))))).. ( -47.50) >DroSim_CAF1 14987 120 - 1 GAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGAC .................((((((((((((.(((((.......))).(((((((((....)))))))...........(((((.(((.....))))))))))))...)))))))))))).. ( -47.50) >DroEre_CAF1 14786 120 - 1 GAUUAUUUUAAGCACGACGUUGUGAUGACGUCUGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGGGCAGCUGGCUCCGCGAACGGUCAUCACAAUGAC .................(((((((((((((((....))).((((....(((((((....)))))))...........(((((.(((.....))))))))))))...)))))))))))).. ( -45.80) >DroYak_CAF1 15157 120 - 1 GAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAGCAUGGAGGAGCAUGGAGCAGCUGGCUCCGCGAACGGUCAUCACAAUGAC .................((((((((((((.(((((....)))(((((((((((((....))))))).....((((......)))).....)))))).....))...)))))))))))).. ( -46.80) >consensus GAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGAGCACGGAGGCGCUGGCUCCGCGAACGGUCAUCACAAUGAC .................(((((((((((((((....))).((((....(((((((....)))))))...........(((((.(((.....))))))))))))...)))))))))))).. (-45.66 = -45.34 + -0.32)

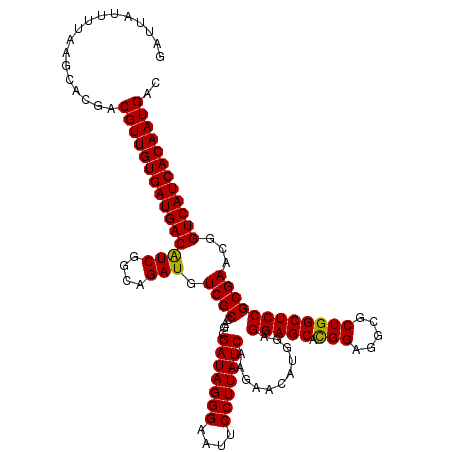

| Location | 18,337,242 – 18,337,362 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18337242 120 + 22224390 UCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGAUACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......)))))).))........(((((...)))))))))))))...))))))............. ( -31.30) >DroSec_CAF1 15193 120 + 1 UCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGAUACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......)))))).))........(((((...)))))))))))))...))))))............. ( -31.30) >DroSim_CAF1 15027 120 + 1 UCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGAUACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......)))))).))........(((((...)))))))))))))...))))))............. ( -31.30) >DroEre_CAF1 14826 120 + 1 UCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCAGACGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGACACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......))))).)))........(((((...)))))))))))))...))))))............. ( -31.50) >DroYak_CAF1 15197 120 + 1 UCCUCCAUGCUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGAUACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......)))))).))........(((((...)))))))))))))...))))))............. ( -31.10) >consensus UCCUCCAUGUUCUUGAUAAGGAAUUCCCUAUCGCUGGCGACAUCUGCCGAUGUCAUCACAACGUCGUGCUUAAAAUAAUCCGUGACGGAUGUGUUGUUCUCCAUGGAUACCUUAAUAUAG ...((((((.....((((.((....)).))))...((((((((..((((((((.......)))))).))........(((((...)))))))))))))...))))))............. (-30.42 = -30.46 + 0.04)

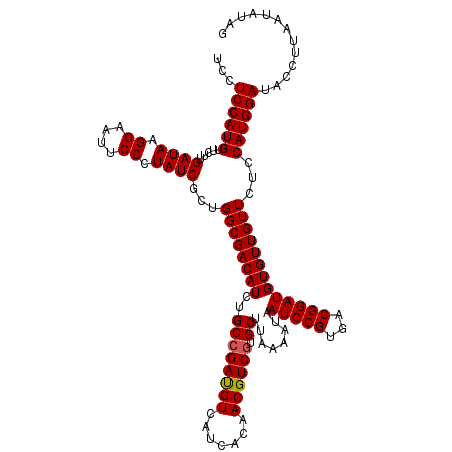
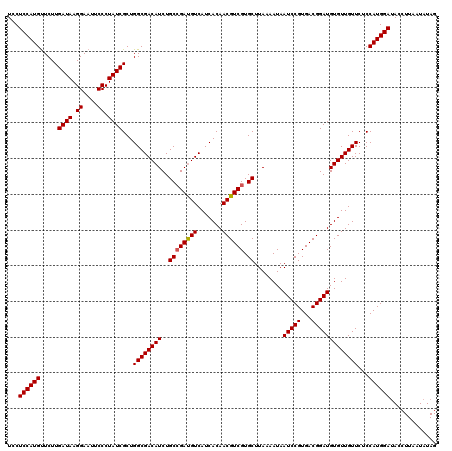
| Location | 18,337,242 – 18,337,362 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18337242 120 - 22224390 CUAUAUUAAGGUAUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGA .............((((((...........(((((...)))))...............((((.(.(((((((....))))))))))))(((((((....))))))).....))))))... ( -34.70) >DroSec_CAF1 15193 120 - 1 CUAUAUUAAGGUAUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGA .............((((((...........(((((...)))))...............((((.(.(((((((....))))))))))))(((((((....))))))).....))))))... ( -34.70) >DroSim_CAF1 15027 120 - 1 CUAUAUUAAGGUAUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGA .............((((((...........(((((...)))))...............((((.(.(((((((....))))))))))))(((((((....))))))).....))))))... ( -34.70) >DroEre_CAF1 14826 120 - 1 CUAUAUUAAGGUGUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACGUCUGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGA ...........(.((((((...........(((((...)))))...............((((.(.(((((((....))))))))))))(((((((....))))))).....)))))).). ( -34.10) >DroYak_CAF1 15197 120 - 1 CUAUAUUAAGGUAUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAGCAUGGAGGA ((((.((((((((((((((((..........)))))...))).))))))))((.....((((.(.(((((((....))))))))))))(((((((....)))))))....)))))).... ( -36.60) >consensus CUAUAUUAAGGUAUCCAUGGAGAACAACACAUCCGUCACGGAUUAUUUUAAGCACGACGUUGUGAUGACAUCGGCAGAUGUCGCCAGCGAUAGGGAAUUCCUUAUCAAGAACAUGGAGGA .............((((((...........(((((...)))))...............((((.(.(((((((....))))))))))))(((((((....))))))).....))))))... (-34.60 = -34.44 + -0.16)

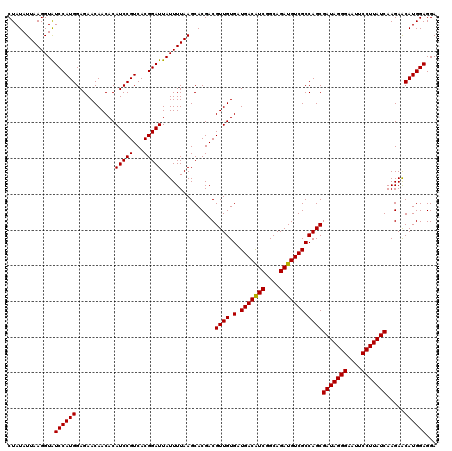
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:39 2006