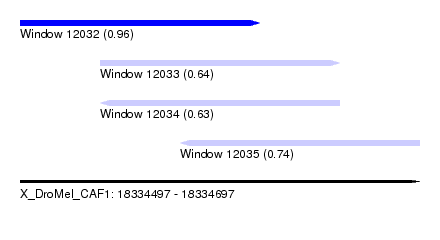
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,334,497 – 18,334,697 |
| Length | 200 |
| Max. P | 0.959729 |

| Location | 18,334,497 – 18,334,617 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18334497 120 + 22224390 GCUACGCGAUCAUCAAUGAUCCUGGCAUACAAUUGGUUGCCAUGGACACACUCAGCGAAUUCUACUGGCUAGAUGAGCCUAAGAUUGCCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAU ((((.(.(((((....)))))))))).......((((...((((((((..(((.((((..(((...((((.....))))..)))))))...)))..))))))))....))))........ ( -36.90) >DroSec_CAF1 12496 120 + 1 GCUACGCGAUCAUCAAUGAUCCUGGCAUACAACUGGUUGCCAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGAUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAU ((((.(.(((((....)))))))))).......((((...((((((((..(((.......(((...((((.....))))..))).......)))..))))))))....))))........ ( -34.34) >DroSim_CAF1 12331 120 + 1 GCUACGCGAUCAUCAAUGAUCCUGGCAUACAACUGGUUGCCAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGCUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAU ((((.(.(((((....)))))))))).......((((...((((((((..((((((.(.......).)))....((((....)))).....)))..))))))))....))))........ ( -32.80) >DroEre_CAF1 12178 120 + 1 GCUAUGCGAUCAUCAAUGAUCCUGCCAUACAACUGGUUGCCAUGGACACACUCAGCGAAUUCUACUGGCUGGACGAGCUUAAGAUAACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAC ((...(((((((....)))))..((((......)))).))((((((((..(((.......(((...((((.....))))..))).......)))..))))))))....)).......... ( -28.94) >DroYak_CAF1 12455 120 + 1 GCUAUGCGAUUAUCAAUGAUCCUGGCAUACAACUGGUGGCCAUGGACACACUCAGCGAAUUCUACUGGCUGGACGACCUAAAGUUAAGCAAGAGAAUGUCCAUGUACCGCCACUAUCGAU ((((.(.(((((....)))))))))).......(((((((((((((((..(((...(....).....(((.(((........))).)))..)))..))))))))....)))))))..... ( -36.00) >consensus GCUACGCGAUCAUCAAUGAUCCUGGCAUACAACUGGUUGCCAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGAUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAU ((((.(.(((((....)))))))))).......((((...((((((((..(((.......(((...((((.....))))..))).......)))..))))))))....))))........ (-29.42 = -29.82 + 0.40)


| Location | 18,334,537 – 18,334,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18334537 120 + 22224390 CAUGGACACACUCAGCGAAUUCUACUGGCUAGAUGAGCCUAAGAUUGCCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAUUGCUCCAGGCGCGCCUGGAGAAGCUCUGCAAGGAUGCCAU ((((((((..(((.((((..(((...((((.....))))..)))))))...)))..)))))))).((((.......))))..(((((((....)))))))..(((((....))).))... ( -40.20) >DroSec_CAF1 12536 120 + 1 CAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGAUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAUUGCUCCAGGCGCGCCUGGAGAAGUUCUGCAAGGACGCCAU ((((((((..(((.......(((...((((.....))))..))).......)))..))))))))....((............(((((((....)))))))..((((.....))))))... ( -39.04) >DroSim_CAF1 12371 120 + 1 CAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGCUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAUUGCUCCAGGCGCGCCUGGAGAAGCUCUGCAAGGACGCCAU ((((((((..((((((.(.......).)))....((((....)))).....)))..))))))))...((((...........(((((((....)))))))..((...))..)).)).... ( -36.20) >DroEre_CAF1 12218 120 + 1 CAUGGACACACUCAGCGAAUUCUACUGGCUGGACGAGCUUAAGAUAACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGACUGCUCCAGGCGCGCCUGGAGAAGCUCUGCAAAGACGCCAU ((((((((..(((.......(((...((((.....))))..))).......)))..))))))))..................(((((((....)))))))..(((((....))).))... ( -36.24) >DroYak_CAF1 12495 120 + 1 CAUGGACACACUCAGCGAAUUCUACUGGCUGGACGACCUAAAGUUAAGCAAGAGAAUGUCCAUGUACCGCCACUAUCGAUUGCUCCUGGCGCGCCUGGAGAAGCUCUGCAAGGACGCCAU ((((((((..(((...(....).....(((.(((........))).)))..)))..))))))))....((((..............))))(((((((.((.....)).).))).)))... ( -33.34) >consensus CAUGGACACACUCAGCGAAUUCUACUGGCUAGACGAGCCUAAGAUUACCAAGAGAAUGUCCAUGUAUCGCCACUAUCGAUUGCUCCAGGCGCGCCUGGAGAAGCUCUGCAAGGACGCCAU ((((((((..(((.......(((...((((.....))))..))).......)))..))))))))..................(((((((....)))))))..(((((....))).))... (-33.00 = -33.24 + 0.24)


| Location | 18,334,537 – 18,334,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -41.84 |
| Consensus MFE | -35.68 |
| Energy contribution | -37.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18334537 120 - 22224390 AUGGCAUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGCAAUCUUAGGCUCAUCUAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG ((.((...((....((....(((((((....)))))))...))...))..)).)).(((((((((...((..(((((((..((((.....))))...)).))).))..)).))))))))) ( -43.30) >DroSec_CAF1 12536 120 - 1 AUGGCGUCCUUGCAGAACUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAUCUUAGGCUCGUCUAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG ....((((((....((....(((((((....)))))))...))...)).))))...(((((((((...((..((..(((..((((.....))))...)))....))..)).))))))))) ( -43.10) >DroSim_CAF1 12371 120 - 1 AUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAGCUUAGGCUCGUCUAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG ....((((((....((....(((((((....)))))))...))...)).))))...(((((((((...((..((...((..((((.....))))...)).....))..)).))))))))) ( -43.30) >DroEre_CAF1 12218 120 - 1 AUGGCGUCUUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAGUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUUAUCUUAAGCUCGUCCAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG ....((((.(((..((.((.(((((((....))))))).))))..))).))))...(((((((((...((..((..(((...(((.....)))....)))....))..)).))))))))) ( -42.00) >DroYak_CAF1 12495 120 - 1 AUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCAGGAGCAAUCGAUAGUGGCGGUACAUGGACAUUCUCUUGCUUAACUUUAGGUCGUCCAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG .((((((.((((.((.....)).)))).))))))...((...(....)..))....(((((((((.(((..((....((...(((......)))...)).....)).))).))))))))) ( -37.50) >consensus AUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAUCUUAGGCUCGUCUAGCCAGUAGAAUUCGCUGAGUGUGUCCAUG ....((((.(((..((....(((((((....)))))))...))..))).))))...(((((((((...((((((..(((..((((.....))))...)))....)))))).))))))))) (-35.68 = -37.12 + 1.44)

| Location | 18,334,577 – 18,334,697 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -36.96 |
| Energy contribution | -38.04 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18334577 120 - 22224390 CUCCAGAAAGGCAGACUCCACCGUGUACAUGCCGCAGACGAUGGCAUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGCAAUCUU .....(..(((.(((.....(((((((..((((((...((((.(((....))).......(((((((....)))))))..))))...)))))).)))))))....))))))..)...... ( -39.60) >DroSec_CAF1 12576 120 - 1 CUCCAGGAAGGCAGACUCCACCGUGUAGAUGCCGCACACGAUGGCGUCCUUGCAGAACUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAUCUU ..((((((.((......)).(((((((..((((((...((((.(((....))).......(((((((....)))))))..))))...)))))).)))))))......))))))....... ( -44.50) >DroSim_CAF1 12411 120 - 1 CUCCAGGAAGGCAGACUCCACCGUGUAGAUGCCGCACACGAUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAGCUU ..((((((.((......)).(((((((..((((((...((((.(((....))).......(((((((....)))))))..))))...)))))).)))))))......))))))....... ( -44.50) >DroEre_CAF1 12258 120 - 1 CUCCAGGAAGGCGGACUCCACCGUGUAGAUUCCGCAGACGAUGGCGUCUUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAGUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUUAUCUU ..((((((.((......)).(((((((....(((((((((....))))).....((.((.(((((((....))))))).))))....))))...)))))))......))))))....... ( -43.30) >DroYak_CAF1 12535 120 - 1 CUCCAGGAACACAGACUCCACCGUGUACAUUCCGCAAACGAUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCAGGAGCAAUCGAUAGUGGCGGUACAUGGACAUUCUCUUGCUUAACUU .....(((........))).((((((((...((((...((((.((.((((.((...(((......)))..)).)))))).))))...))))..))))))))................... ( -38.10) >consensus CUCCAGGAAGGCAGACUCCACCGUGUAGAUGCCGCACACGAUGGCGUCCUUGCAGAGCUUCUCCAGGCGCGCCUGGAGCAAUCGAUAGUGGCGAUACAUGGACAUUCUCUUGGUAAUCUU ..((((((.((......)).(((((((..((((((...((((.(((....))).......(((((((....)))))))..))))...)))))).)))))))......))))))....... (-36.96 = -38.04 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:35 2006