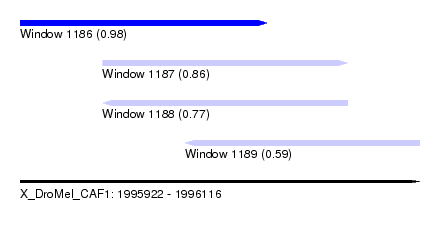
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,995,922 – 1,996,116 |
| Length | 194 |
| Max. P | 0.983933 |

| Location | 1,995,922 – 1,996,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.03 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1995922 120 + 22224390 AACCCACCCCUUUCCACCAUUUUCCGUGGCCUGUACAAACCGAUUUCCCAAACAAACAUUAUACAAUUACCCUCAAAAGGGGAUCCAUUCUGAUGUCCCAAGGGGUGUACACGUAAUCCC ....((((((((.((((........))))..........................((((((........((((....)))).........))))))...))))))))............. ( -24.53) >DroSec_CAF1 27915 118 + 1 AACCCACCCCUUUCCACCAUUUUCCGUGGCCUGUACAAACCGAUUUCCCAAACAAACAUUAUACAAUUUCCCUCAAAAGGGGAUCCAUUCUGAGUUCCCAAGGGGUGUA--UGUAAUCCC ....((((((((.((((........)))).................................................(((((..(.....)..)))))))))))))..--......... ( -24.90) >DroSim_CAF1 22120 118 + 1 AACCCACCCCUUUCCACCAUUUUCCGUGGCCUGUACAAACCGAUUUCCCAAACAAACGUUAUACAAUUACCCUCAAAAGGAGAUCCAUUCUGAGUUCCCAAGGGGUGUA--UGUAAUCCC ....((((((((.((((........))))............(((((((..............................)))))))..............))))))))..--......... ( -22.31) >DroEre_CAF1 17698 118 + 1 AACCCACCCUCUUCCACCAUUUUCCGUGGCCUGUGCAAACCGAUUUCCCAAGCAAACAUUAAACAAUUACCCUCAAAAGGAGAUCCAUUCCGAUGUCCCAAGGGGUGUG--CGGAAUACC ..(((((((((((((((........))))....(((...............)))........................(((.(((......))).))).)))))).)))--.))...... ( -23.06) >DroYak_CAF1 28527 118 + 1 AACCCACCCUCUUCCACCAUUUUCCGUGGUCCGUACAAACCGAUUUCCCAAACAAACAUUAAACAAUUACCCUCAAAAGGGGAUCCAUUCCGAUGUCCCAAGGGGUGCA--CGGAAUCCC .....................((((((((((.((....)).))).......................((((((.....(((((((......))).))))..))))))))--))))).... ( -23.30) >consensus AACCCACCCCUUUCCACCAUUUUCCGUGGCCUGUACAAACCGAUUUCCCAAACAAACAUUAUACAAUUACCCUCAAAAGGGGAUCCAUUCUGAUGUCCCAAGGGGUGUA__CGUAAUCCC ....((((((((.((((........))))............(((((((..............................)))))))..............))))))))............. (-20.11 = -20.03 + -0.08)

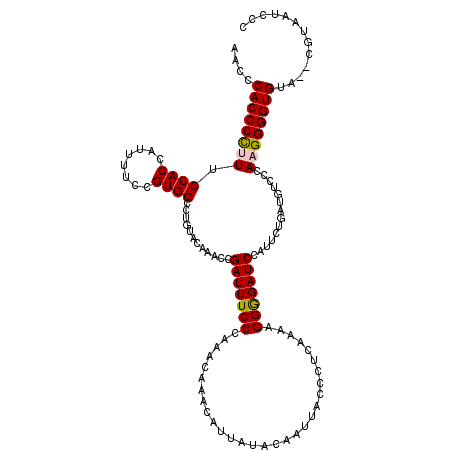
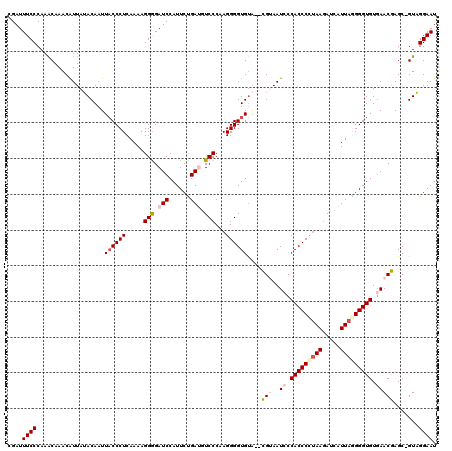
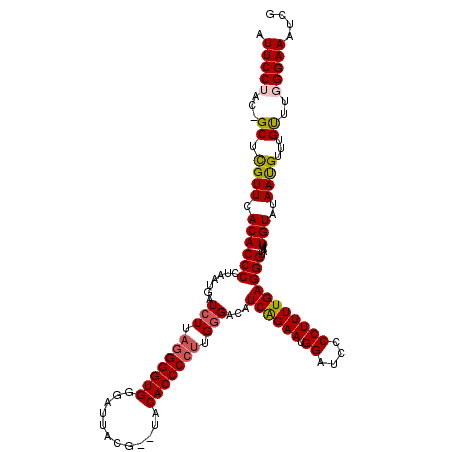
| Location | 1,995,962 – 1,996,081 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -24.88 |
| Energy contribution | -26.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1995962 119 + 22224390 CGAUUUCCCAAACAAACAUUAUACAAUUACCCUCAAAAGGGGAUCCAUUCUGAUGUCCCAAGGGGUGUACACGUAAUCCCACCCCUAAGAUCAUUAGGGGUGUGAACGAGC-GUAGGAAU ....((((.............((((....((((.....(((((((......))).)))).)))).)))).((((..((.(((((((((.....))))))))).))....))-)).)))). ( -35.00) >DroSec_CAF1 27955 117 + 1 CGAUUUCCCAAACAAACAUUAUACAAUUUCCCUCAAAAGGGGAUCCAUUCUGAGUUCCCAAGGGGUGUA--UGUAAUCCCACCCCUAACAUCAUUAAGGGUGUGAACAAGC-GUAGGAAU ...........................((((((....))))))(((((.((..((((...(((((((..--........))))))).(((((......))))))))).)).-)).))).. ( -28.30) >DroSim_CAF1 22160 117 + 1 CGAUUUCCCAAACAAACGUUAUACAAUUACCCUCAAAAGGAGAUCCAUUCUGAGUUCCCAAGGGGUGUA--UGUAAUCCCACCCCUAAGAUCAUUAAGGGUGUGAACAAGC-GUAGGAAU ....((((.......(((((.((((..((((((.....((((.((......)).))))...))))))..--)))).((.(((((.(((.....))).))))).))...)))-)).)))). ( -28.30) >DroEre_CAF1 17738 118 + 1 CGAUUUCCCAAGCAAACAUUAAACAAUUACCCUCAAAAGGAGAUCCAUUCCGAUGUCCCAAGGGGUGUG--CGGAAUACCACCCUGAAGAUCAUUAGGGGUGUGAACGAGCCGUUGGAAU ....((((...................((((((.....(((.(((......))).)))...)))))).(--(((..(((..((((((......))))))..)))......)))).)))). ( -29.20) >DroYak_CAF1 28567 117 + 1 CGAUUUCCCAAACAAACAUUAAACAAUUACCCUCAAAAGGGGAUCCAUUCCGAUGUCCCAAGGGGUGCA--CGGAAUCCCACCCCUAAGAUCAUUAGGGGUGUAAACGAGC-GUUGGAAU .............................((((....)))).....(((((((((..(...(((((...--....)))))((((((((.....))))))))......)..)-)))))))) ( -35.90) >consensus CGAUUUCCCAAACAAACAUUAUACAAUUACCCUCAAAAGGGGAUCCAUUCUGAUGUCCCAAGGGGUGUA__CGUAAUCCCACCCCUAAGAUCAUUAGGGGUGUGAACGAGC_GUAGGAAU ....((((...................((((((.....(((.(((......))).)))...))))))....(((..((.(((((((((.....))))))))).))))).......)))). (-24.88 = -26.08 + 1.20)

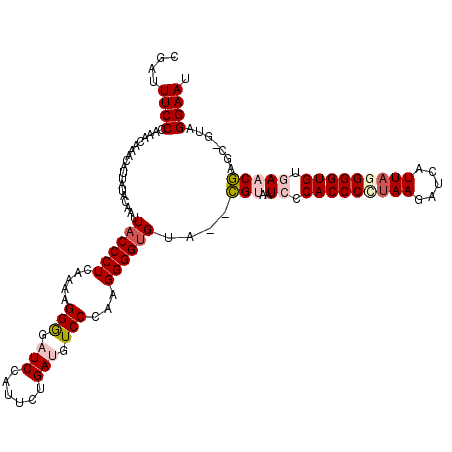
| Location | 1,995,962 – 1,996,081 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -27.84 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1995962 119 - 22224390 AUUCCUAC-GCUCGUUCACACCCCUAAUGAUCUUAGGGGUGGGAUUACGUGUACACCCCUUGGGACAUCAGAAUGGAUCCCCUUUUGAGGGUAAUUGUAUAAUGUUUGUUUGGGAAAUCG .((((((.-((..((((.(((((((((.....))))))))))))).(((((((((.(((((((((..((......)))))).....)))))....))))).))))..)).)))))).... ( -38.60) >DroSec_CAF1 27955 117 - 1 AUUCCUAC-GCUUGUUCACACCCUUAAUGAUGUUAGGGGUGGGAUUACA--UACACCCCUUGGGAACUCAGAAUGGAUCCCCUUUUGAGGGAAAUUGUAUAAUGUUUGUUUGGGAAAUCG .((((((.-((..((((.((((((((((...))))))))))))))...(--(((((((...)))..(((((((.((....)))))))))......))))).......)).)))))).... ( -36.20) >DroSim_CAF1 22160 117 - 1 AUUCCUAC-GCUUGUUCACACCCUUAAUGAUCUUAGGGGUGGGAUUACA--UACACCCCUUGGGAACUCAGAAUGGAUCUCCUUUUGAGGGUAAUUGUAUAACGUUUGUUUGGGAAAUCG .((((((.-((.((((..(((((((((.....))))))))).((((((.--....(((...)))..(((((((.((....))))))))).))))))....))))...)).)))))).... ( -37.40) >DroEre_CAF1 17738 118 - 1 AUUCCAACGGCUCGUUCACACCCCUAAUGAUCUUCAGGGUGGUAUUCCG--CACACCCCUUGGGACAUCGGAAUGGAUCUCCUUUUGAGGGUAAUUGUUUAAUGUUUGCUUGGGAAAUCG .((((...(((.((((.(((.((((.(((.(((...(((((((.....)--).)))))...))).)))(((((.((....)))))))))))....)))..))))...)))..)))).... ( -29.10) >DroYak_CAF1 28567 117 - 1 AUUCCAAC-GCUCGUUUACACCCCUAAUGAUCUUAGGGGUGGGAUUCCG--UGCACCCCUUGGGACAUCGGAAUGGAUCCCCUUUUGAGGGUAAUUGUUUAAUGUUUGUUUGGGAAAUCG .(((((((-(..((((..(((((((((.....)))))))))..((((((--((..((....))..)).)))))).....((((....)))).........))))..)).))))))..... ( -37.20) >consensus AUUCCUAC_GCUCGUUCACACCCCUAAUGAUCUUAGGGGUGGGAUUACG__UACACCCCUUGGGACAUCAGAAUGGAUCCCCUUUUGAGGGUAAUUGUAUAAUGUUUGUUUGGGAAAUCG .(((((...((.((((.((((((.......(((.(((((((............))))))).)))...((((((.((....)))))))))))....)))..))))...))..))))).... (-27.84 = -28.04 + 0.20)

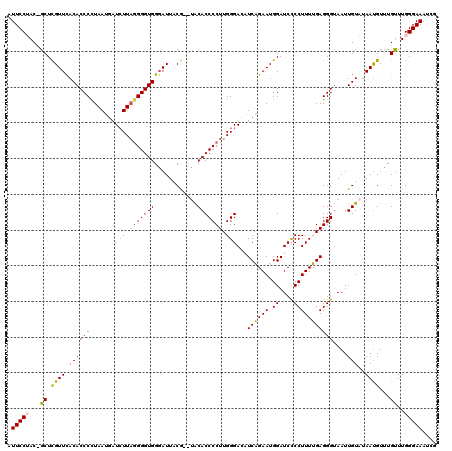
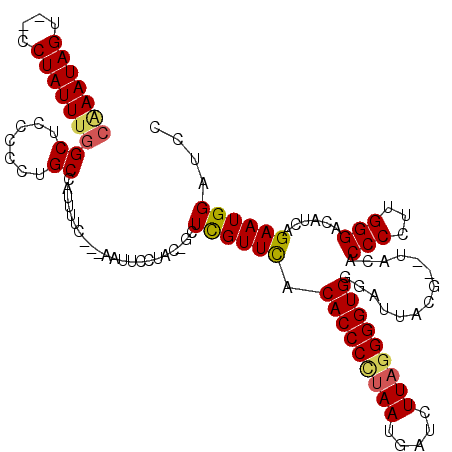
| Location | 1,996,002 – 1,996,116 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1996002 114 - 22224390 CAAAUAGU--CCUAUUUGGCUCCCCCUGCCAUUUUC---AAUUCCUAC-GCUCGUUCACACCCCUAAUGAUCUUAGGGGUGGGAUUACGUGUACACCCCUUGGGACAUCAGAAUGGAUCC ......((--((((..((((.......)))).....---......(((-((..((((.(((((((((.....)))))))))))))...))))).......)))))).............. ( -35.20) >DroSec_CAF1 27995 112 - 1 CAAAUAGU--CCUAUUUGGCUCCCCCUGCCAUUUUC---AAUUCCUAC-GCUUGUUCACACCCUUAAUGAUGUUAGGGGUGGGAUUACA--UACACCCCUUGGGAACUCAGAAUGGAUCC ((((((..--..))))))..........(((((((.---(.((((((.-....((((.((((((((((...))))))))))))))....--.........)))))).).))))))).... ( -27.97) >DroSim_CAF1 22200 112 - 1 CAAAUAGU--CCUAUUUGGCUCCCCCUGCCAUUUUC---AAUUCCUAC-GCUUGUUCACACCCUUAAUGAUCUUAGGGGUGGGAUUACA--UACACCCCUUGGGAACUCAGAAUGGAUCU ((((((..--..))))))..........(((((((.---(.((((((.-....((((.(((((((((.....)))))))))))))....--.........)))))).).))))))).... ( -27.47) >DroEre_CAF1 17778 116 - 1 CGAAUAGU--CCUAUUUGGCUCCCGCUGCAUUUUUCAGGAAUUCCAACGGCUCGUUCACACCCCUAAUGAUCUUCAGGGUGGUAUUCCG--CACACCCCUUGGGACAUCGGAAUGGAUCU .(((((((--(......(((....)))..........((....))...)))).)))).(((((....((.....))))))).(((((((--....(((...)))....)))))))..... ( -27.30) >DroYak_CAF1 28607 114 - 1 --AAUAGUCUCCUAUUUGGCUCGC-CUGCAUUUUUCAGAAAUUCCAAC-GCUCGUUUACACCCCUAAUGAUCUUAGGGGUGGGAUUCCG--UGCACCCCUUGGGACAUCGGAAUGGAUCC --.......(((......((....-..))...........(((((...-((.((.(..(((((((((.....)))))))))..)...))--.)).(((...))).....))))))))... ( -31.80) >consensus CAAAUAGU__CCUAUUUGGCUCCCCCUGCCAUUUUC___AAUUCCUAC_GCUCGUUCACACCCCUAAUGAUCUUAGGGGUGGGAUUACG__UACACCCCUUGGGACAUCAGAAUGGAUCC (((((((....)))))))((.......))......................((((((.(((((((((.....)))))))))..............(((...)))......)))))).... (-23.46 = -23.58 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:29 2006