
| Sequence ID | X_DroMel_CAF1 |
|---|---|
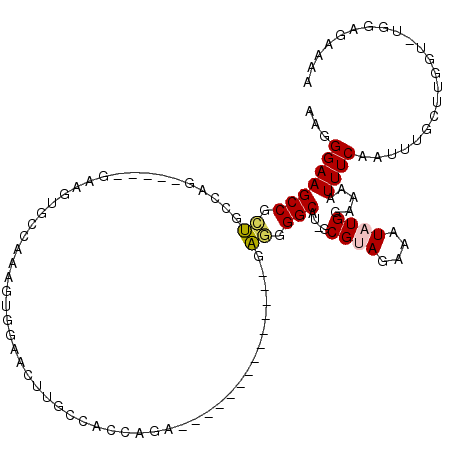
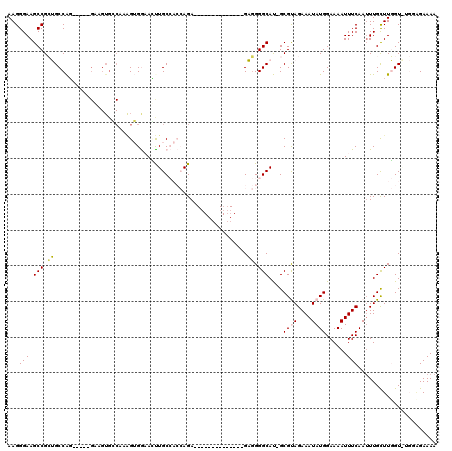
| Location | 18,290,748 – 18,290,851 |
| Length | 103 |
| Max. P | 0.992854 |

| Location | 18,290,748 – 18,290,851 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.04 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -6.70 |
| Energy contribution | -7.02 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18290748 103 + 22224390 AAGGGAAGCCGCUGCCAG-----GAGGUGGCAAGGUGGAAUCUGCCACCAGAA-----------ACGAGGGGCAU-GCGUAGAAAUAUGGAAAAUUUCAAUUUGCUUGGUAUGGAGAAAA ........(((.((((((-----(.(((((((.(((...))))))))))....-----------..((((....(-.((((....)))).)...))))......))))))))))...... ( -30.30) >DroPse_CAF1 1229 98 + 1 AAGGGAAGCCGAAGCCGCGUCUGGCAGUGCCACAGGGACAGUUGC----AGA--------------GUGGGGCACAGCGCAGAAAUAUGGAAAAUUUCAAUUUGUUUUGU----GGAAAA ..((....))...(((.((((((.(((((((....)).)).))))----)))--------------.)).)))....(((((((...((((....)))).....))))))----)..... ( -25.40) >DroSec_CAF1 3731 103 + 1 AAGGGAAGCCGCUGCCAG-----GAGGUGGCAAGGUGGAACCUGCCACCAGAA-----------UCGAGGGGCAU-GCGUAGAAAUAUGGAAAAUUUCAAUUUGCUUGGUAUGGAGAAAA ........(((.((((((-----(.(((((((.((.....))))))))).(((-----------(.((((....(-.((((....)))).)...)))).)))).))))))))))...... ( -33.90) >DroYak_CAF1 4016 103 + 1 AAGGGAAGCCGCUGCCAG----------------GUGGAACUUGCCACCAGACGACGCACGACGAGGAGGGGCAU-GCGUAGAAAUAUGGAAAAUUUCAAAUUGCUUGGUGUGGUGAAAA .......(((((.(((((----------------(..(....((((.((...(..((.....)).)..))))))(-.((((....)))).)..........)..)))))))))))..... ( -28.90) >DroPer_CAF1 1281 98 + 1 AAGGGAAGCCGAAGCCGCGUCUGGCAGUGCCACAGGGACAGUUGC----AGA--------------GUUGGGCACAGCGCAGAAAUAUGGAAAAUUUCAAUUUGUUUUGU----GGAAAA ..((....))...((((....))))....(((((((.((((.(((----...--------------((((....)))))))(((((.......)))))...)))))))))----)).... ( -23.90) >consensus AAGGGAAGCCGCUGCCAG_____GAAGUGCCAAAGUGGAACUUGCCACCAGA______________GAGGGGCAU_GCGUAGAAAUAUGGAAAAUUUCAAUUUGCUUGGU_UGGAGAAAA ..((....))...((((.........................((((........................))))..(((..(((((.......)))))....))).)))).......... ( -6.70 = -7.02 + 0.32)

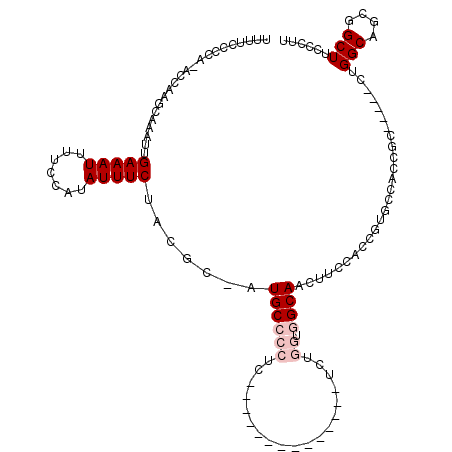
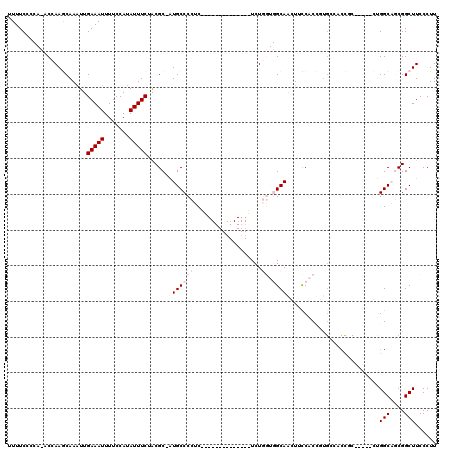
| Location | 18,290,748 – 18,290,851 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.04 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -4.91 |
| Energy contribution | -6.11 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.20 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18290748 103 - 22224390 UUUUCUCCAUACCAAGCAAAUUGAAAUUUUCCAUAUUUCUACGC-AUGCCCCUCGU-----------UUCUGGUGGCAGAUUCCACCUUGCCACCUC-----CUGGCAGCGGCUUCCCUU .............((((.....(((((.......)))))..(((-.((((......-----------....((((((((........))))))))..-----..)))))))))))..... ( -26.24) >DroPse_CAF1 1229 98 - 1 UUUUCC----ACAAAACAAAUUGAAAUUUUCCAUAUUUCUGCGCUGUGCCCCAC--------------UCU----GCAACUGUCCCUGUGGCACUGCCAGACGCGGCUUCGGCUUCCCUU ......----...........................((((.((.(((((.((.--------------...----((....))...)).))))).)))))).(.(((....))).).... ( -21.20) >DroSec_CAF1 3731 103 - 1 UUUUCUCCAUACCAAGCAAAUUGAAAUUUUCCAUAUUUCUACGC-AUGCCCCUCGA-----------UUCUGGUGGCAGGUUCCACCUUGCCACCUC-----CUGGCAGCGGCUUCCCUU .............((((.....(((((.......)))))..(((-.((((....((-----------....(((((((((......)))))))))))-----..)))))))))))..... ( -29.50) >DroYak_CAF1 4016 103 - 1 UUUUCACCACACCAAGCAAUUUGAAAUUUUCCAUAUUUCUACGC-AUGCCCCUCCUCGUCGUGCGUCGUCUGGUGGCAAGUUCCAC----------------CUGGCAGCGGCUUCCCUU ......((.(((((..(.....(((((.......))))).((((-((((........).))))))).)..)))))....(((((..----------------..)).)))))........ ( -23.80) >DroPer_CAF1 1281 98 - 1 UUUUCC----ACAAAACAAAUUGAAAUUUUCCAUAUUUCUGCGCUGUGCCCAAC--------------UCU----GCAACUGUCCCUGUGGCACUGCCAGACGCGGCUUCGGCUUCCCUU ......----...........................((((.((.(((((((..--------------...----((....))...)).))))).)))))).(.(((....))).).... ( -20.60) >consensus UUUUCCCCA_ACCAAGCAAAUUGAAAUUUUCCAUAUUUCUACGC_AUGCCCCUC______________UCUGGUGGCAACUUCCACCGUGCCACCGC_____CUGGCAGCGGCUUCCCUU ......................(((((.......))))).......((((((...................)).))))..........................(((....)))...... ( -4.91 = -6.11 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:16 2006