
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,288,929 – 18,289,055 |
| Length | 126 |
| Max. P | 0.888325 |

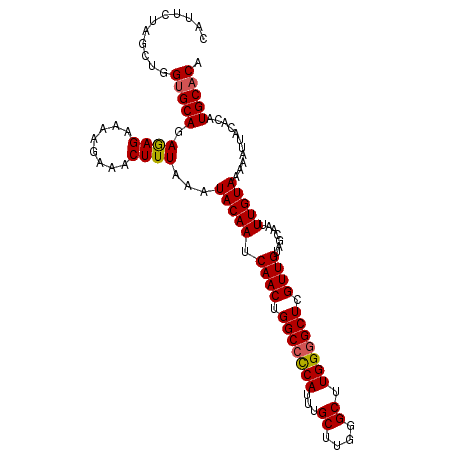
| Location | 18,288,929 – 18,289,031 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18288929 102 - 22224390 CAUUCUAUCUGGUGCAGAGAG-----AAACUUUAAAUACAAUCAACUGGCCUCAUUUGCUUGGGCUUGGCGCUCGUUGUAGCAAUUUGUAAAAAUUACACAUGCACA ..((((.((((...)))).))-----))..............((((.(((.(((...((....)).))).))).))))..(((...((((.....))))..)))... ( -19.60) >DroSec_CAF1 1938 107 - 1 CAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUAGCAAUUUGUAAAAAUUACACAUGCACA ...........(((((.((((........))))...(((((.((((.(((((((...((....)).))))))).)))).......)))))...........))))). ( -29.30) >DroSim_CAF1 2021 107 - 1 CAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUAGCAAUUUGUAAAAAUUACACAUGCACA ...........(((((.((((........))))...(((((.((((.(((((((...((....)).))))))).)))).......)))))...........))))). ( -29.30) >DroYak_CAF1 2101 107 - 1 CAUUUUAUCUGGCGCAGAAAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCCUUGGCUUGGGGCUCGUUGUAGCAAUUUGUAAAAAUUACACAUGCACA ..((((.((((...)))).))))...................((((.(((((((...((....)).))))))).))))..(((...((((.....))))..)))... ( -27.60) >consensus CAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUAGCAAUUUGUAAAAAUUACACAUGCACA ...........(((((.((((........))))...(((((.((((.(((((((...((....)).))))))).)))).......)))))...........))))). (-25.98 = -26.10 + 0.12)


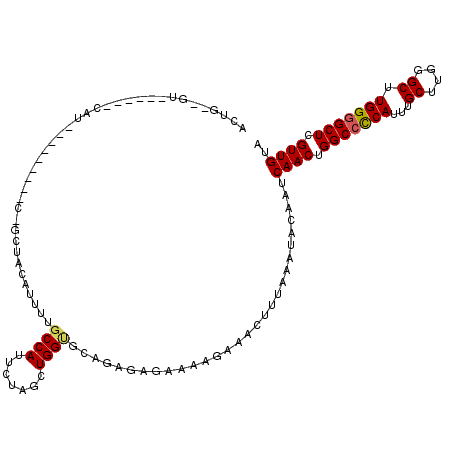
| Location | 18,288,956 – 18,289,055 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.82 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18288956 99 - 22224390 ACUG--GU-----ACAU---------UUGCUACAUUUUGCCAUUCUAUCUGGUGCAGAGAG-----AAACUUUAAAUACAAUCAACUGGCCUCAUUUGCUUGGGCUUGGCGCUCGUUGUA ..((--((-----(...---------.)))))......((((((((.((((...)))).))-----))...................((((.((......)))))))))).......... ( -18.60) >DroSec_CAF1 1965 113 - 1 ACUG--GU-----UCGUCCGCCAACCCAGCUACUUUUUGCCAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUA .((.--.(-----((.((.((....(((((((.............))))))).)).))..))).))................((((.(((((((...((....)).))))))).)))).. ( -33.22) >DroSim_CAF1 2048 113 - 1 NNNN--NN-----NNNNNNNNNNNNNNNNNNNNNNNNNNCCAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUA ....--..-----.............................((((..(((...)))..))))...................((((.(((((((...((....)).))))))).)))).. ( -24.70) >DroYak_CAF1 2128 120 - 1 ACUUCGGUCUUCCCCACCAAAAAACCCCGCUCCAUUUUGCCAUUUUAUCUGGCGCAGAAAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCCUUGGCUUGGGGCUCGUUGUA ..(((..(((((..(.(((.........((........)).........))).)..).))))...)))..............((((.(((((((...((....)).))))))).)))).. ( -28.67) >consensus ACUG__GU______CAU_________C_GCUACAUUUUGCCAUUCUAGCUGGUGCAGAGAGAAAAGAAACUUUAAAUACAAUCAACUGGCCCCAUUUGCUUGGGCUUGGGGCUCGUUGUA ......................................((((.......)))).............................((((.(((((((...((....)).))))))).)))).. (-20.29 = -20.48 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:15 2006