
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,287,199 – 18,287,558 |
| Length | 359 |
| Max. P | 0.977301 |

| Location | 18,287,199 – 18,287,310 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -22.83 |
| Energy contribution | -24.14 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287199 111 + 22224390 CCACACC-CCCAGAUUCUUGCCACCUGCCCCGGCCCACAUUAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUUGCUGCUGUUGCUGCUUGGCGGGAAAAUCGGUGGAAGAAUG .......-.....((((((.(((((.((....))((........--------..(((((.....)))))(((((((.((.((....)).)).)).)))))))......))))))))))). ( -38.40) >DroSec_CAF1 329 97 + 1 -----CC-CCCUGAUUCUUGCCACCUGCCCUGCCCCACAUUAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUG -----..-.....((((((.(((.(..(((((((..........--------..(((((.....))))).((.(((....---------)))))..))))))).....).))))))))). ( -33.10) >DroSim_CAF1 368 97 + 1 -----CC-CCCUGAUUCUUGCCACCUGCCCUGCCCCACAUUAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUG -----..-.....((((((.(((.(..(((((((..........--------..(((((.....))))).((.(((....---------)))))..))))))).....).))))))))). ( -33.10) >DroYak_CAF1 381 104 + 1 CCACACCACCCUGAUUCUUGCCACCUGC-----CCCACAUUAUCGGUGGCACGAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUCGCCGGCAAAAUCG--GGCAGAGUG ...((((..(((((((.(((((...(((-----(((........)).))))((((((((.....))))).((.(((....---------))))))))..))))))))))--))..).))) ( -38.70) >consensus _____CC_CCCUGAUUCUUGCCACCUGCCCUGCCCCACAUUAUC________AAUGGCAUUAGUUGCCACGCCAGCUGUU_________GCUGCUUGGCGGGAAAAUCGCUGGAAGAAUG .............((((((.(((.........((((..................(((((.....))))).((((((.((..........)).))).))))))).......))))))))). (-22.83 = -24.14 + 1.31)



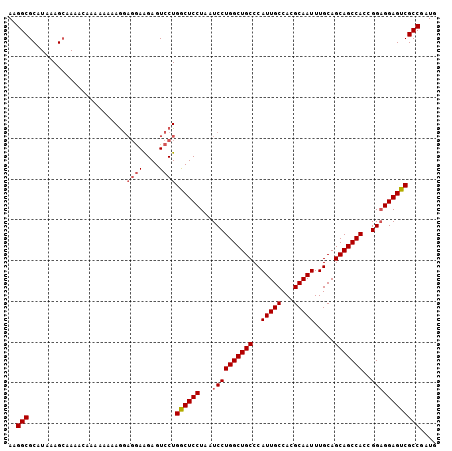
| Location | 18,287,238 – 18,287,348 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -25.73 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287238 110 + 22224390 UAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUUGCUGCUGUUGCUGCUUGGCGGGAAAAUCGGUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCG--GGCGAUUGC ((((--------(.(((((.....)))))((((..((.(..(((...((.((((...)))).))...)))..).))....))))..)))))......(((((((.....--))))))).. ( -35.70) >DroSec_CAF1 363 101 + 1 UAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCG--GGCGAUUGC ((((--------(.(((((.....)))))..(((.(((((---------.((..(..((((.....))))..).))))))).))).)))))......(((((((.....--))))))).. ( -34.90) >DroSim_CAF1 402 101 + 1 UAUC--------AAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUGGGUGAGCGAUAAAAAUGGAUUGCCUUGCG--GGCGAUUGC ((((--------..(((((.....))))).((((.(((((---------.((..(..((((.....))))..).))))))).)).))))))......(((((((.....--))))))).. ( -34.60) >DroYak_CAF1 416 109 + 1 UAUCGGUGGCACGAUGGCAUUAGUUGCCACGCCAGCUGUU---------GCUGCUCGCCGGCAAAAUCG--GGCAGAGUGGGUGGGUGAUAAAAAUUGAUUGCCUAACGGGGGCGAUUGC .((((......))))((((.....)))).((((..(((((---------.(..((((((.(......).--))).)))..)..(((..((........))..)))))))).))))..... ( -39.60) >consensus UAUC________AAUGGCAUUAGUUGCCACGCCAGCUGUU_________GCUGCUUGGCGGGAAAAUCGCUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCG__GGCGAUUGC ..............(((((.....)))))(((((((.((..........)).))).))))....(((((((....(...(((..(.((((....)))).)..)))..)...))))))).. (-25.73 = -26.60 + 0.87)

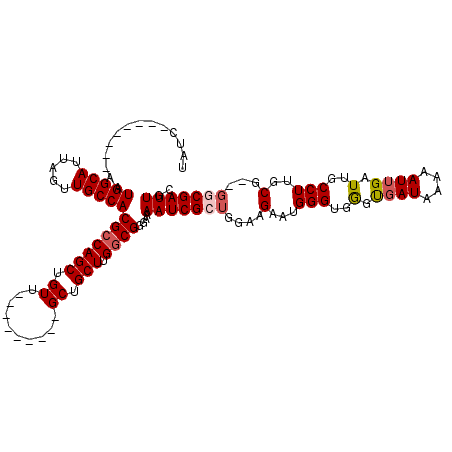
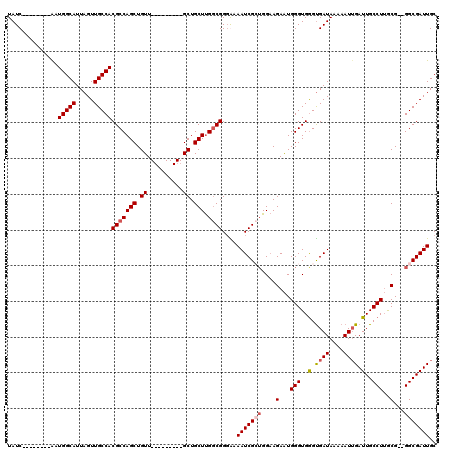
| Location | 18,287,270 – 18,287,376 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287270 106 + 22224390 GCUGCUGUUGCUGCUUGGCGGGAAAAUCGGUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAAAAA .....((((..(((((.((.....(((((((.......................)))))))(((((((((....)))))))))))...))))).))))........ ( -27.90) >DroSec_CAF1 395 97 + 1 ---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAGGAG ---------..(((((.((......((((((.(.........).))))))...........(((((((((....)))))))))))...)))))............. ( -26.90) >DroSim_CAF1 434 93 + 1 ---------GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUGGGUGAGCGAUAAAAAUGGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGAAAA----AAAUAAG ---------....(((.((......((((((.(.........).))))))...........(((((((((....)))))))))))...)))....----....... ( -24.20) >consensus _________GCUGCUUGGCGGGGAAAUCGCUGGAAGAAUGGGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAAAAG ...........(((((.((......((((((.(.........).))))))...........(((((((((....)))))))))))...)))))............. (-23.94 = -24.17 + 0.23)

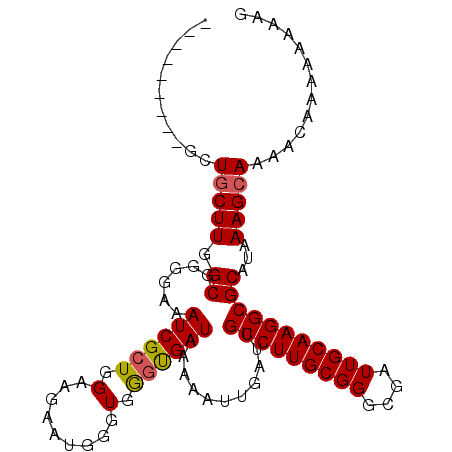

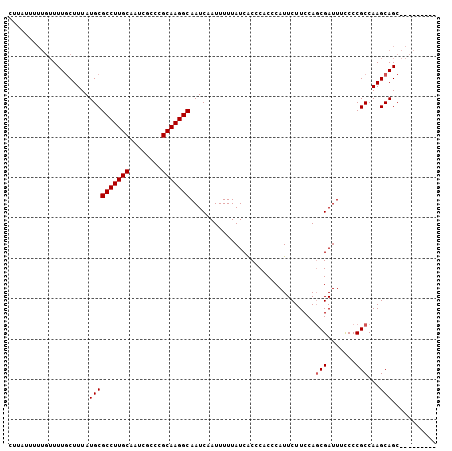
| Location | 18,287,270 – 18,287,376 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287270 106 - 22224390 UUUUUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACCCAUUCUUCCACCGAUUUUCCCGCCAAGCAGCAACAGCAGC .......(((((((((((...(((((((((........)))))))........................((.......)).......)).)))))).))))).... ( -21.30) >DroSec_CAF1 395 97 - 1 CUCCUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACCCAUUCUUCCAGCGAUUUCCCCGCCAAGCAGC--------- ............((((((.....(((((((........)))))))...............................(((.......))).)))))).--------- ( -20.10) >DroSim_CAF1 434 93 - 1 CUUAUUU----UUUUCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCCAUUUUUAUCGCUCACCCAUUCUUCCAGCGAUUUCCCCGCCAAGCAGC--------- .......----.........((((((((((........)))))))...........((((((.............))))))...........)))..--------- ( -21.02) >consensus CUUAUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACCCAUUCUUCCAGCGAUUUCCCCGCCAAGCAGC_________ ....................((((((((((........)))))))...............................(((.......)))...)))........... (-16.93 = -17.27 + 0.33)

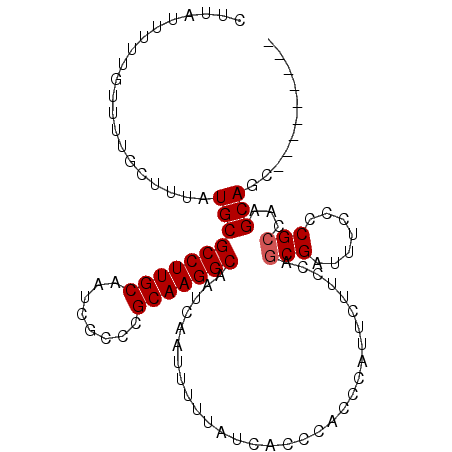
| Location | 18,287,310 – 18,287,410 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.37 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287310 100 + 22224390 GGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAAAAA---AAGGAGUCCUGACUCCUAAUCCUGGCUGCCCAUU .((((((........(((...(((((((((....)))))))))((.....))....))).......---.((((((....))))))..........)))))). ( -32.60) >DroSec_CAF1 426 103 + 1 GGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAGGAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUU .((((((........(((...(((((((((....)))))))))((.....))....)))..((((..((((...((....))))))..))))....)))))). ( -34.90) >DroSim_CAF1 465 99 + 1 GGUGAGCGAUAAAAAUGGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGAAAA----AAAUAAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUU ((..(((.(.......((((((((((((((....)))))))))............----.....(((((..((....))..))))))))))).)))..))... ( -30.91) >consensus GGUGGGUGAUAAAAAUUGAUUGCCUUGCGGGCGAUUGCAAGGCGCAUAAAGCAAAACAAAAAAAAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUU .((((((..............(((((((((....))))))))).....................((((....((((....))))....))))....)))))). (-26.37 = -27.27 + 0.89)


| Location | 18,287,310 – 18,287,410 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287310 100 - 22224390 AAUGGGCAGCCAGGAUUAGGAGUCAGGACUCCUU---UUUUUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACC ..(((((((..((((..((((((....)))))).---.))))..)))....((.....))(((((((........)))))))...............)))).. ( -29.80) >DroSec_CAF1 426 103 - 1 AAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUCCUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACC ..((((.(((.(((((.(((((..((((....)))).)))))....))))))))......(((((((........)))))))...............)))).. ( -31.60) >DroSim_CAF1 465 99 - 1 AAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUUAUUU----UUUUCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCCAUUUUUAUCGCUCACC ..(((((....((((..(((((......)))))..)))).....----............(((((((........)))))))..............))))).. ( -28.30) >consensus AAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUUAUUUUUGUUUUGCUUUAUGCGCCUUGCAAUCGCCCGCAAGGCAAUCAAUUUUUAUCACCCACC ..((((.....((((..(((((......)))))..)))).....................(((((((........)))))))...............)))).. (-24.06 = -24.07 + 0.01)



| Location | 18,287,348 – 18,287,450 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -30.39 |
| Energy contribution | -31.50 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287348 102 + 22224390 AAGGCGCAUAAAGCAAAACAAAAAAAAA---AAGGAGUCCUGACUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUG ..(((((.....))..............---..........(((((((.....(((((((..(((((...))))).....)))))))....)))))))))).... ( -34.10) >DroSec_CAF1 464 105 + 1 AAGGCGCAUAAAGCAAAACAAAAAGGAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGCGGAGUCGCCGAUG ..(((((.....)).........((((..((((...((....))))))..)))).(((((..(((((...)))))..))))).)))..(((((....)))))... ( -38.20) >DroSim_CAF1 503 101 + 1 AAGGCGCAUAAAGAAAA----AAAUAAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUG ..(((............----........((((....))))(((((((.....(((((((..(((((...))))).....)))))))....)))))))))).... ( -35.30) >consensus AAGGCGCAUAAAGCAAAACAAAAAAAAGGAGGAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUG ..(((........................((((....))))((((((...((((((((((..(((((...))))).....)))))))..)))))))))))).... (-30.39 = -31.50 + 1.11)


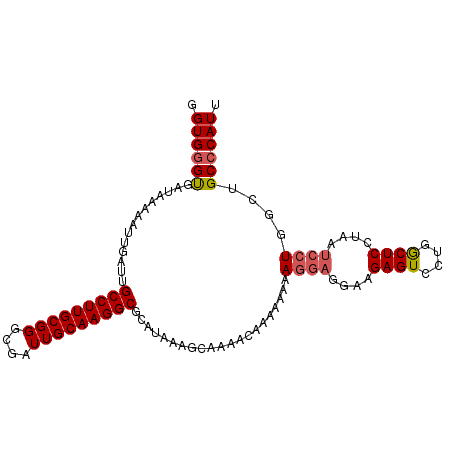
| Location | 18,287,348 – 18,287,450 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287348 102 - 22224390 CAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGCAAUGGGCAGCCAGGAUUAGGAGUCAGGACUCCUU---UUUUUUUUUGUUUUGCUUUAUGCGCCUU ....((((((((((((..((((((((....(((((...))))).)))))))))))...))))))..........---..............((.....))))).. ( -36.90) >DroSec_CAF1 464 105 - 1 CAUCGGCGACUCCGCCGGUGGCUGCUGCAAAUUGCGUGGCAAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUCCUUUUUGUUUUGCUUUAUGCGCCUU .(((((((....)))))))(((.(((((..(((((...)))))..)))))((((((.(((((..((((....)))).)))))....))))))........))).. ( -41.50) >DroSim_CAF1 503 101 - 1 CAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGCAAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUUAUUU----UUUUCUUUAUGCGCCUU ....((((.(((((((..((((((((....(((((...))))).)))))))))))...))))..((((....))))........----...........)))).. ( -33.70) >consensus CAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGCAAUGGGCAGCCAGGAUUAGGAGCCAGGACUCUUCCUCCUUAUUUUUGUUUUGCUUUAUGCGCCUU ....((((..........((((((((....(((((...))))).))))))))(((..(((((......)))))..))).....................)))).. (-30.44 = -30.67 + 0.23)

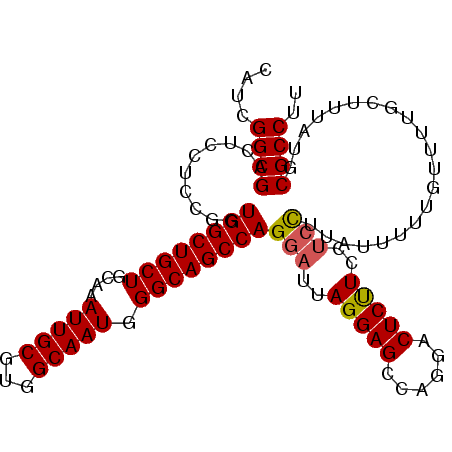

| Location | 18,287,376 – 18,287,490 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -41.02 |
| Consensus MFE | -32.65 |
| Energy contribution | -32.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287376 114 + 22224390 ------AAGGAGUCCUGACUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUGGGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAA ------..(((((...(((((((.....(((((((..(((((...))))).....)))))))....)))))))...(((((.((.((....)).)).)))))...))))).......... ( -40.70) >DroSec_CAF1 492 117 + 1 --GAG-GAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGCGGAGUCGCCGAUGGGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAA --..(-(((......(((((..((((..(((((((..(((((...)))))..))))).(((..(((((....)))))...)))..))..))))..)))))......)))).......... ( -40.90) >DroSim_CAF1 527 117 + 1 --GAG-GAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUGGGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAA --..(-(((.......(((((((.....(((((((..(((((...))))).....)))))))....)))))))...(((((.((.((....)).)).)))))....)))).......... ( -39.70) >DroYak_CAF1 565 120 + 1 GGGAGUAAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCGGGCAGCGGAGGAGUCGCCGAUGGGCAACAUCAUUGCUGGCCAUUAAAAUUGCGUUAAACGAA ((((.....((((....))))....))))(((........)))(((((((((.....((.((((((.(..((.(((....))).))..).)))))).))....)))))))))........ ( -42.80) >consensus __GAG_AAAGAGUCCUGGCUCCUAAUCCUGGCUGCCCAUUGCCACGCAAUUUGCAGCAGCCACCGGAGGAGUCGCCGAUGGGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAA ................((((((...((((((((((..(((((...))))).....)))))))..)))))))))...(((((.((.((....)).)).))))).......((.....)).. (-32.65 = -32.77 + 0.12)



| Location | 18,287,410 – 18,287,518 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287410 108 - 22224390 GAUAUGCACU----------UCGCAUUUUCAGUUU--UGUUUCGUUUAACGGAAUUUUAAUGGCCAGCAAUGAUGUUGCCCAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGC ((.((((...----------..))))..))(((((--((((......)))))))))......(((((((((..(((.(((((((((.(....).)))))))..)).))).))))).)))) ( -34.50) >DroSec_CAF1 529 108 - 1 GAUAUGCACU----------UCGCAUUUUCAGUUU--UGUUUCGUUUAACGGAAUUUUAAUGGCCAGCAAUGAUGUUGCCCAUCGGCGACUCCGCCGGUGGCUGCUGCAAAUUGCGUGGC ((.((((...----------..))))..))(((((--((((......)))))))))......(((((((((..(((.(((((((((((....)))))))))..)).))).))))).)))) ( -40.30) >DroSim_CAF1 564 108 - 1 GAUAUGCACU----------UCGCAUUUUCAGUUU--UGUUUCGUUUAACGGAAUUUUAAUGGCCAGCAAUGAUGUUGCCCAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGC ((.((((...----------..))))..))(((((--((((......)))))))))......(((((((((..(((.(((((((((.(....).)))))))..)).))).))))).)))) ( -34.50) >DroYak_CAF1 605 120 - 1 GACUUGCACUGCAAGUUGAUUCGCAUUUUCAGUUGUGUGUUUCGUUUAACGCAAUUUUAAUGGCCAGCAAUGAUGUUGCCCAUCGGCGACUCCUCCGCUGCCCGCUGCAAAUUGCGUGGC (((((((...)))))))((..(((((........)))))..)).....(((((((((((.(((.((((...(..((((((....))))))..)...)))).))).)).)))))))))... ( -41.60) >consensus GAUAUGCACU__________UCGCAUUUUCAGUUU__UGUUUCGUUUAACGGAAUUUUAAUGGCCAGCAAUGAUGUUGCCCAUCGGCGACUCCUCCGGUGGCUGCUGCAAAUUGCGUGGC .....................((((.(((((((.....((((((.....))))))......(((((.(...(..((((((....))))))..)...).))))))))).))).)))).... (-28.16 = -28.48 + 0.31)

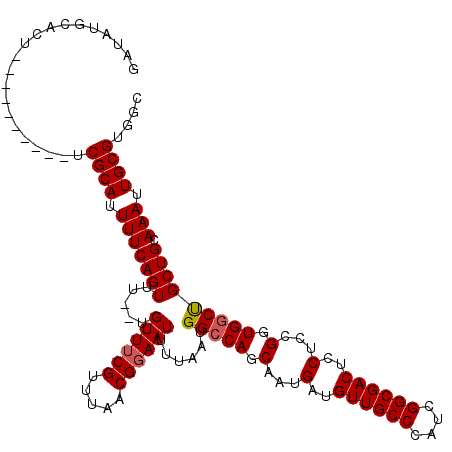
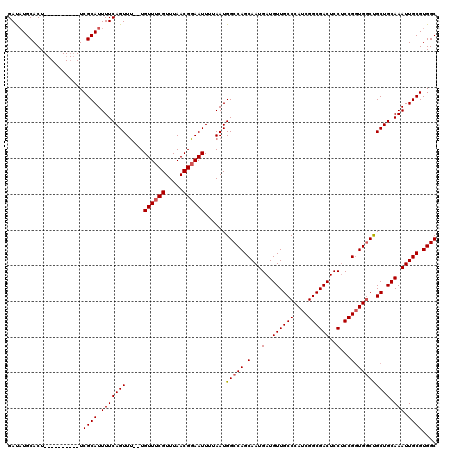
| Location | 18,287,450 – 18,287,558 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.62 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18287450 108 + 22224390 GGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAAACA--AAACUGAAAAUGCGA----------AGUGCAUAUCGCACGAUUCGCCAAUAAGGAUUUUAUGCGCCAGCAUGUAA (....).....((((((((((.(((((((.(((......))).--...........((((----------.((((.....))))...))))......)))))))))).)))))))..... ( -31.40) >DroSec_CAF1 569 108 + 1 GGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAAACA--AAACUGAAAAUGCGA----------AGUGCAUAUCGCACGAUUCGCCAAUAAGGAUUUUAUGCGCCACCAUGUAA (((((.....)))))((((((.(((((((.(((......))).--...........((((----------.((((.....))))...))))......)))))))))).)))......... ( -25.30) >DroSim_CAF1 604 108 + 1 GGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAAACA--AAACUGAAAAUGCGA----------AGUGCAUAUCGCACGAUUCGCCAAUAAGGAUUUUAUGCGCCACCAUGUAA (((((.....)))))((((((.(((((((.(((......))).--...........((((----------.((((.....))))...))))......)))))))))).)))......... ( -25.30) >DroYak_CAF1 645 120 + 1 GGCAACAUCAUUGCUGGCCAUUAAAAUUGCGUUAAACGAAACACACAACUGAAAAUGCGAAUCAACUUGCAGUGCAAGUCUCACGAUUCGCCAAUAAGACUUUUAUGUCCCAACUUGUAA (((..((....))...))).........(.(((......))).)((((........(((((((.((((((...)))))).....)))))))......(((......))).....)))).. ( -22.20) >consensus GGCAACAUCAUUGCUGGCCAUUAAAAUUCCGUUAAACGAAACA__AAACUGAAAAUGCGA__________AGUGCAUAUCGCACGAUUCGCCAAUAAGGAUUUUAUGCGCCACCAUGUAA (((((.....)))))((((((.(((((((.(((......)))..............((((...........((((.....))))...))))......)))))))))).)))......... (-17.60 = -18.41 + 0.81)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:13 2006