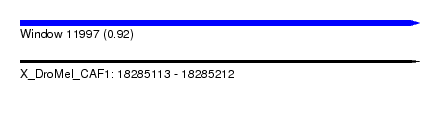
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,285,113 – 18,285,212 |
| Length | 99 |
| Max. P | 0.919297 |

| Location | 18,285,113 – 18,285,212 |
|---|---|
| Length | 99 |
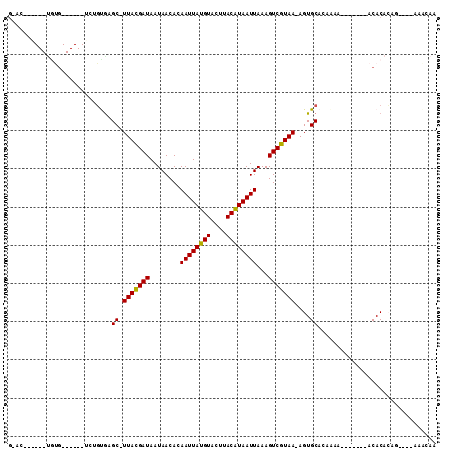
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
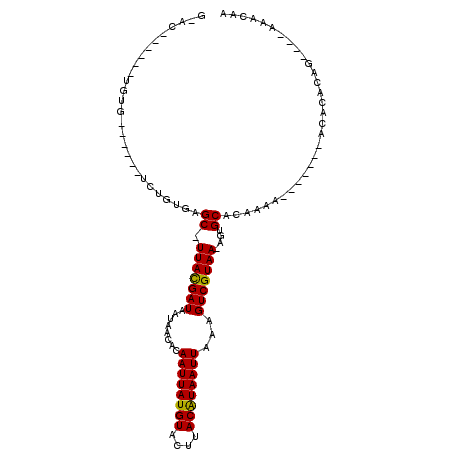
| Mean single sequence MFE | -17.48 |
| Consensus MFE | -10.28 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18285113 99 + 22224390 GGGCCGAGCAUGUGUUGCGCUCUGCGGGC-UUACGAUAAUAACACAAUUAUGUACUUACGUAAUUAAAGUCGUAA-AGUGCACAAAA-------ACACACAG----AAACAA ..........(((((((((((.(((((.(-((..(.......)..((((((((....)))))))).)))))))).-)))))).....-------)))))...----...... ( -20.50) >DroPse_CAF1 5189 106 + 1 GUAGACGGCAUGUGUUGCGCUCUGUGAGC-UUACGAUAAUAACACAAUUAUGUACUUACAUAAUUAAAGUCGUAA-AGUGCAAAAAAAAAACACACACACAG----AAACAA .......(((.....)))..((((((...-(((((((........((((((((....))))))))...)))))))-.(((...........)))...)))))----)..... ( -20.70) >DroGri_CAF1 10563 92 + 1 GGAC------UGUG------UCCGUACGC-UUAUGAUAAUAACACAAUUAUGUACUUACAUAAUUAAAGUCGUAA-AGUGCACAAAAA------ACACAAACUAAGAAGCAA ((((------...)------)))((((..-(((((((........((((((((....))))))))...)))))))-.)))).......------.................. ( -17.00) >DroEre_CAF1 4741 76 + 1 G----------------------GCGGGC-UUACGAUAAUAACACAAUUAUGUACUUACGUAAUUAAAGUCGUAA-AGUGCACAA-A-------ACACACAG----AAACAA .----------------------(.(.((-(((((((........((((((((....))))))))...))))).)-))).).)..-.-------........----...... ( -12.10) >DroMoj_CAF1 6880 88 + 1 GCAC------CAAG------UCCUUGCGC-UUAUGAUAAUAACACAAUUAUGUACUUACAUAAUUAAAGUCGUAAAAGUGCACAAA--------ACACAGAC---AAAACAA ....------....------....(((((-(((((((........((((((((....))))))))...))))))..))))))....--------........---....... ( -13.80) >DroPer_CAF1 5243 104 + 1 NNNNNNGGCAUGUGUUGCGCUCUGUGAGCCUUACGAUAAUAACACAAUUAUGUACUUACAUAAUUAAAGUCGUAA-AGUGCAAAGAAAA---ACACACACAG----AAACAA .......(((.....)))..((((((....(((((((........((((((((....))))))))...)))))))-.(((.........---.))).)))))----)..... ( -20.80) >consensus G_AC______UGUG______UCUGUGAGC_UUACGAUAAUAACACAAUUAUGUACUUACAUAAUUAAAGUCGUAA_AGUGCACAAAA_______ACACACAG____AAACAA ...........................((.(((((((........((((((((....))))))))...)))))))....))............................... (-10.28 = -9.83 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:02 2006