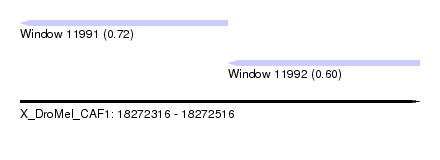
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,272,316 – 18,272,516 |
| Length | 200 |
| Max. P | 0.717530 |

| Location | 18,272,316 – 18,272,420 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.47 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18272316 104 - 22224390 UUUGGGUCACCUUGGUGAUAUGUUUAGUUGAGCUCACAAGCAUUUGUAGUUUAAAGACGGAGUACACGUUUUUUUUGACUCUAAUGCCGUUUGGGGUCACCUUU ...(((((((....))))).........(((.((((...(((((.(.((((.(((((((.......)))))))...))))).)))))....)))).)))))... ( -28.40) >DroSec_CAF1 40618 88 - 1 UUUGGAUCACCUUGGUGAUAUGUUUAAUAU---------GC-----CACUUUAAAGUCGGAGUGCAAGUUU--UUUGACUCUGAUGCUGUUUGGGGUCACCUUG ...(((((((....)))))..........(---------((-----.(((((......)))))))).....--..((((((..(......)..))))))))... ( -21.90) >DroSim_CAF1 15253 88 - 1 UUUGGGUCACCUUGGUGAUAUGUUUAAUAC---------GC-----CACUUUAAAGUCGGAGUGCAAGUUU--UUUGACUCUGAUGCUGUUUGGGGUCACCUUG ...((((.(((((((((.(((.....))))---------))-----)........((((((((.((((...--)))))))))))).......))))).)))).. ( -25.60) >consensus UUUGGGUCACCUUGGUGAUAUGUUUAAUAC_________GC_____CACUUUAAAGUCGGAGUGCAAGUUU__UUUGACUCUGAUGCUGUUUGGGGUCACCUUG ...(((((((....)))))...........................((((((......))))))...........(((((((((......)))))))))))... (-19.02 = -18.47 + -0.55)


| Location | 18,272,420 – 18,272,516 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -18.65 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18272420 96 - 22224390 GGGGGUGACUCUUCGGCGGUGACCUUUUUAAAGAGCAACAAAUAAGGUUGAGUUCUUAAAGGUAUGCCACUUUAAAAUGGUAACCGGAUUCCAAGU--------------- ((((....))))..((((((.(((.((((((((.(((((...(((((......)))))...)).)))..)))))))).))).))))....))....--------------- ( -27.90) >DroSec_CAF1 40706 109 - 1 GG-GGUGACUCGUUAGCGGUGACCUUUUCAAAGAGCUACAGAAAAGUUUGAGUUCUUAAAGGUAUGCCACUUUAGAAUGGUACCC-GAUUCCAAGUUAUGGGUCACCUAGU .(-((((((((((.(((((((((((((...(((((((.((((....))))))))))))))))).))))......((((.(....)-.))))...))))))))))))))... ( -39.10) >DroSim_CAF1 15341 109 - 1 GG-GGUAACUCUUUAGCGGUGACCUUUUCAAAGAGCUUCAGAAAAGUUUGAGUUCUUAAAGGUAUGCCACUUUAAAAUGGUACCC-GAUUCCAAGUUAUGGGUCACCUAGU .(-(((.((((..((((((((((((((...(((((((.((((....))))))))))))))))).)))).........(((.....-....))).)))).)))).))))... ( -31.20) >consensus GG_GGUGACUCUUUAGCGGUGACCUUUUCAAAGAGCUACAGAAAAGUUUGAGUUCUUAAAGGUAUGCCACUUUAAAAUGGUACCC_GAUUCCAAGUUAUGGGUCACCUAGU ....((((((....((.((((((((((...(((((((.((((....))))))))))))))))).)))).))......(((..........)))))))))............ (-18.65 = -19.77 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:58 2006