| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,271,455 – 18,271,564 |
| Length | 109 |
| Max. P | 0.917593 |

| Location | 18,271,455 – 18,271,564 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -52.70 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
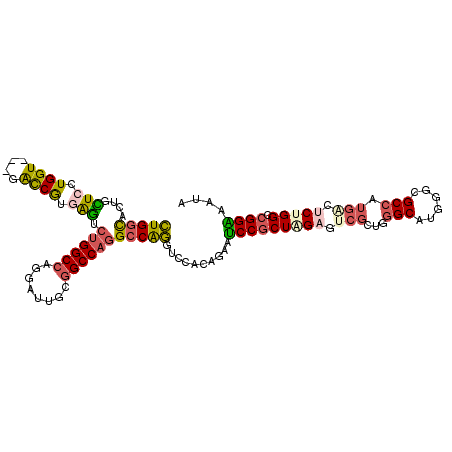
>X_DroMel_CAF1 18271455 109 + 22224390 CUGGCACUUCUCCUGGU---UGUCGCGGGUCUGGCCAUGAUUGCGGCCAGGCCAGGUCCACAGAAUCCGCUAGAGUCGCUGGGCAUGGGCGCCAUGUCUCUGGCCGGAAAUA ..((((((......)).---))))(.((..((((((.((........))))))))..)).)....((((((((((.((.(((((....)).))))).)))))).)))).... ( -45.80) >DroSec_CAF1 39765 109 + 1 CUGGCACUUUUCCUGGU---UAUCGUGGGUCUGGCCAUGAUUGCGGCCAGGCCAGGUCCACAGAAUCCGCUAGAGUCGCUGGGCAUGGGCGCCAUGACUCUGGCCGGAAAUA ........(((((.((.---..((((((..((((((.((........))))))))..)))).))..))((((((((((.(((((....)).))))))))))))).))))).. ( -52.50) >DroSim_CAF1 14401 109 + 1 CUGGCCCUUCUCCUGGU---UAUCGUGGGUCUGGCCAUGAUUGCGGCCAGGCCAGGUCCACAGAAUCCGCUAGAGUCGCUGGGCAUGGGCGCCAUGACUCUGGCCGGAAAUA .(((((........)))---))..((((..((((((.((........))))))))..))))....(((((((((((((.(((((....)).)))))))))))).)))).... ( -52.60) >DroEre_CAF1 44152 109 + 1 CUGGCACUGCUCCUGGU---GGCCGUGAGUCUGGCCAGGAUCGCGGCCAGGCCAGGGCCACAGAAUCCGCUCGAGUCGCUGGGCAUAGGCGCGAUGAGUCUGGCCGGGAAUA (((((...(((((((.(---((((.((.((((((((........)))))))))).)))))))).((((((..(.(((....))).)..))).)))))))...)))))..... ( -54.80) >DroYak_CAF1 36315 109 + 1 CUGGCACUGCUACUGGU---GGCCGUGAGUCUGGCCAGGAUUGCGGCCAGGCCAGGUGCACAGAAUCCGCUCGAAUCGCUGGGCAUGGGCGCCAUGGCGCUGGCCGGGAAUA (((((..(((((.((((---(.(((((..(((((((.((.......)).)))))))..))).......(((((......)))))..)).))))))))))...)))))..... ( -51.60) >DroAna_CAF1 48082 112 + 1 UUGAUUGUGAUACUGGUACUGACCACCAUCCCGGCGGGGAUCGUGGCCAAGCCGAGGGCCCAGAACCCCCUGGAGAACCUCGGCAUGACGGCCAUGAACCUGGCCGGGAACA .............((((....))))...(((((((.(((.((((((((..(((((((..((((......))))....))))))).....)))))))).))).)))))))... ( -58.90) >consensus CUGGCACUGCUCCUGGU___GACCGUGAGUCUGGCCAGGAUUGCGGCCAGGCCAGGUCCACAGAAUCCGCUAGAGUCGCUGGGCAUGGGCGCCAUGACUCUGGCCGGAAAUA (((((....(((.((((....)))).))).((((((........)))))))))))..........(((((((((.(((...(((......))).))).))))).)))).... (-32.20 = -32.48 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:56 2006