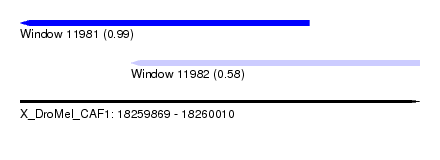
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,259,869 – 18,260,010 |
| Length | 141 |
| Max. P | 0.985303 |

| Location | 18,259,869 – 18,259,971 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18259869 102 - 22224390 ACUUAAAGGUUGUCAAUGGCAACCACCUGAGAUGGCCUGACCA-CCACCACCA------CCACCCACUGGACCGGUGGCACAACAUUCCUGCACAGGGCC-AAAAUUCAU .((((..((((((.....))))))...)))).(((((((.(((-((....(((------........)))...)))))))........((....))))))-)........ ( -28.70) >DroSec_CAF1 33683 99 - 1 ACUUAAAGGUUGUCAACGGCAACCACCUGACAUGGCCUGACCA-CCAC---CA------CCACCCCCUAGGCCGGUGGCACAAGAUUCCUGCACAGGGUC-AAAAUCCAU .......((((((.....)))))).......((((..(((((.-((((---(.------((........))..)))))..........((....))))))-)....)))) ( -27.30) >DroSim_CAF1 8231 99 - 1 ACUUAAAGGUUGUCAACGGCAACCACCUGAGAUGGCCUGACCA-CCAG---CA------CCACCCACUAGGCCGGUGGCACAAGAUUCCUGCACAGGGCC-AAAAUCCAU .((((..((((((.....))))))...)))).(((((((.(((-((.(---(.------...........)).))))))...((....)).....)))))-)........ ( -29.20) >DroEre_CAF1 36836 94 - 1 ACUUAAAGGUUGUCAACGGCAACCACCUGACCUGGCCUAUCCA----------------CCACCCACUAGGCCGGUGGCACAACAUUAUUGCCCAGGCCCUAAAGCACAU .......((((((.....)))))).(((((((.((((((....----------------........)))))))))((((.........))))))))............. ( -29.30) >DroYak_CAF1 30045 98 - 1 ACUUAAAGGUUGUCAACGGCAACCACCUGACAUGGCCUAACCAACCAC---CAACUUACCCACCCACUUG----GUG----AACAUUGCUGCACAGGGCC-AAAAUACAU .......((((((.....))))))........((((((.......(((---(((.............)))----)))----.....((.....)))))))-)........ ( -23.22) >consensus ACUUAAAGGUUGUCAACGGCAACCACCUGACAUGGCCUGACCA_CCAC___CA______CCACCCACUAGGCCGGUGGCACAACAUUCCUGCACAGGGCC_AAAAUACAU .......((((((.....)))))).((((...(((.....)))................(((((.........)))))...............))))............. (-19.50 = -19.90 + 0.40)

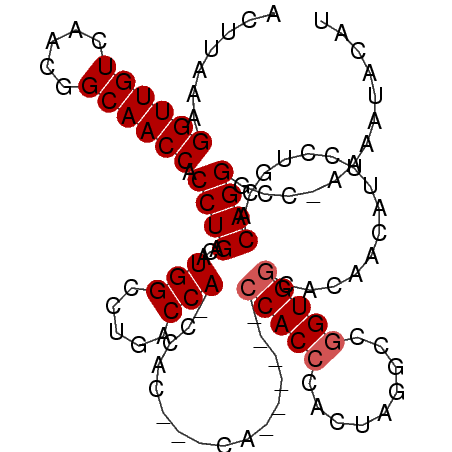
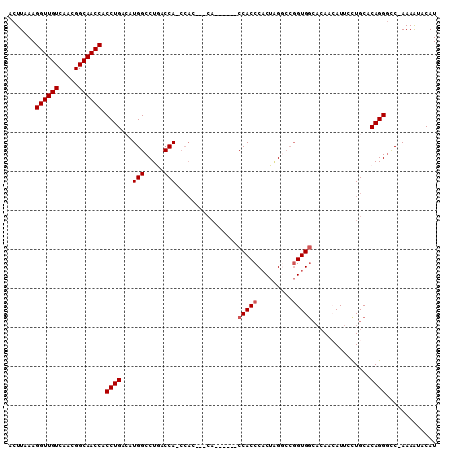
| Location | 18,259,908 – 18,260,010 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.48 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -10.87 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18259908 102 - 22224390 AUAC-ACCAGCCAGCCCCAGGGCUGCUCGCCCAAUGUGCCACUUAAAGGUUGUCAAUGGCAACCACCUGAGAUG-GCCUGACCA-CCACCACCA------CCACCCACUGG ....-.((((.(((((....)))))............((((((((..((((((.....))))))...)))).))-)).......-.........------.......)))) ( -29.70) >DroSec_CAF1 33722 99 - 1 AUAC-ACCAGCCAGCCCCAGGGCUCCCCGUCCAAUGCGCCACUUAAAGGUUGUCAACGGCAACCACCUGACAUG-GCCUGACCA-CCAC---CA------CCACCCCCUAG ....-..((((((....((((((............))..........((((((.....)))))).))))...))-).)))....-....---..------........... ( -22.40) >DroSim_CAF1 8270 99 - 1 AUAC-ACCAGCCAGCCCGAGGGCUCCCCGCCCAAUGCGCCACUUAAAGGUUGUCAACGGCAACCACCUGAGAUG-GCCUGACCA-CCAG---CA------CCACCCACUAG ....-..(((...((....((((.....))))...))((((((((..((((((.....))))))...)))).))-)))))....-....---..------........... ( -26.90) >DroEre_CAF1 36876 79 - 1 ACAC-A--AGCU---GC---------CCGCCCCAUGCGCCACUUAAAGGUUGUCAACGGCAACCACCUGACCUG-GCCUAUCCA----------------CCACCCACUAG ....-.--....---((---------((((.....))).........((((((.....)))))).........)-)).......----------------........... ( -14.40) >DroYak_CAF1 30076 101 - 1 AUAC-ACCAGCC---UCCGGGGCU--CCGCCCAAUGCGGCACUUAAAGGUUGUCAACGGCAACCACCUGACAUG-GCCUAACCAACCAC---CAACUUACCCACCCACUUG ....-....(((---..((((...--((((.....))))........((((((.....)))))).))))....)-))............---................... ( -23.20) >DroAna_CAF1 39401 91 - 1 AUUCUACGAGUCC-UUCGAGAACGGACGACCCCAC---ACACUUAACGGUUGUCAGCGGCAACCACCUGUCACAAGACGGGGGG----------------UCACCCACGAG ......((.((((-.........))))((((((..---.........((((((.....))))))..(((((....)))))))))----------------)).....)).. ( -34.10) >consensus AUAC_ACCAGCCA_CCCCAGGGCU_CCCGCCCAAUGCGCCACUUAAAGGUUGUCAACGGCAACCACCUGACAUG_GCCUGACCA_CCAC___CA______CCACCCACUAG .......(((.........((((.....))))...............((((((.....))))))..))).......................................... (-10.87 = -12.28 + 1.42)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:49 2006