| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 18,242,651 – 18,242,762 |
| Length | 111 |
| Max. P | 0.632794 |

| Location | 18,242,651 – 18,242,762 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -27.51 |
| Energy contribution | -29.32 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18242651 111 + 22224390 UUUGGCUGCCCUGGCCGUGUUUGGGUAUUUCGCCCGAUUUUAGUGACAGUCGGCGAGAGUCGUUAACGAUUGCCACUUGGGCAAUGUGAUUUGGAUAUCUAAAGGUGUCUG ......(((((((((..((((......(((((((.((((........)))))))))))......))))...))))...))))).........(((((((....))))))). ( -35.10) >DroSec_CAF1 16793 111 + 1 CUUGGUUGCCCUGGCCCUGUUUGGGUAUUUCGCCCGAUUUCAGUGACAGUCGGCGAGAGUCGUUAACGAUUGCCACUUGGGUGGUGUGAUUUGAAUAUCUAAAGGUGUCUG ............((((((..(((((((((((((((.(((..((((.((((((((....)))(....)))))).))))..))))).))))....)))))))))))).))).. ( -33.00) >DroEre_CAF1 15981 108 + 1 UAUGGCCGCCCUGGC--UGUU-GAGGAUUUCGCCCGAUUUCAGUGGAAGUUGGCGACUCGCGUUAACGAUUGCCACUUGGGCGGUGUGAUUUGGAUAUCUAAAGGUGUCUG ....(((((((((((--((((-((.(...(((((.((((((....)))))))))))....).))))))...))))...))))))).......(((((((....))))))). ( -43.50) >DroYak_CAF1 16903 105 + 1 UAUGGCUGCCCUGGC--UGUU-UGGGAUUUCGCCCGAUUUCAGUGG---CUGUCGACUCGCGUUAACGAUUGCCACUUGGGCGGUGUGGUUUGGAUAGCUAAAGGUGUCUG ...(((.(((.((((--((((-..((...(((((((.....(((((---(.((((...........)))).))))))))))))).....))..))))))))..)))))).. ( -41.30) >consensus UAUGGCUGCCCUGGC__UGUU_GGGGAUUUCGCCCGAUUUCAGUGACAGUCGGCGACACGCGUUAACGAUUGCCACUUGGGCGGUGUGAUUUGGAUAUCUAAAGGUGUCUG ....(((((((((((..((((......(((((((.((((........)))))))))))......))))...))))...))))))).......(((((((....))))))). (-27.51 = -29.32 + 1.81)

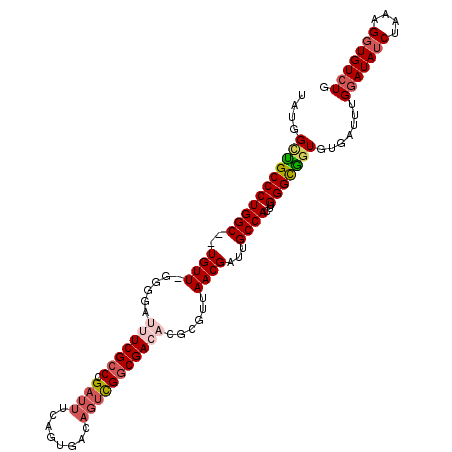

| Location | 18,242,651 – 18,242,762 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 18242651 111 - 22224390 CAGACACCUUUAGAUAUCCAAAUCACAUUGCCCAAGUGGCAAUCGUUAACGACUCUCGCCGACUGUCACUAAAAUCGGGCGAAAUACCCAAACACGGCCAGGGCAGCCAAA ............(((......)))...((((((.(((((((.(((....((.....)).))).)))))))......((.((.............)).)).))))))..... ( -26.62) >DroSec_CAF1 16793 111 - 1 CAGACACCUUUAGAUAUUCAAAUCACACCACCCAAGUGGCAAUCGUUAACGACUCUCGCCGACUGUCACUGAAAUCGGGCGAAAUACCCAAACAGGGCCAGGGCAACCAAG ............(((......)))......(((.(((((((.(((....((.....)).))).)))))))......(((.......))).....)))...((....))... ( -25.30) >DroEre_CAF1 15981 108 - 1 CAGACACCUUUAGAUAUCCAAAUCACACCGCCCAAGUGGCAAUCGUUAACGCGAGUCGCCAACUUCCACUGAAAUCGGGCGAAAUCCUC-AACA--GCCAGGGCGGCCAUA ............(((......)))...((((((...((((..((((....)))).(((((...(((....)))....))))).......-....--))))))))))..... ( -30.10) >DroYak_CAF1 16903 105 - 1 CAGACACCUUUAGCUAUCCAAACCACACCGCCCAAGUGGCAAUCGUUAACGCGAGUCGACAG---CCACUGAAAUCGGGCGAAAUCCCA-AACA--GCCAGGGCAGCCAUA ............(((.............(((((.((((((....((..((....))..)).)---)))))(....))))))........-....--((....))))).... ( -28.70) >consensus CAGACACCUUUAGAUAUCCAAAUCACACCGCCCAAGUGGCAAUCGUUAACGACACUCGCCAACUGCCACUGAAAUCGGGCGAAAUACCC_AACA__GCCAGGGCAGCCAAA ...........................((((((...((((....(((........(((((.................)))))........)))...))))))))))..... (-17.44 = -18.45 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:47 2006